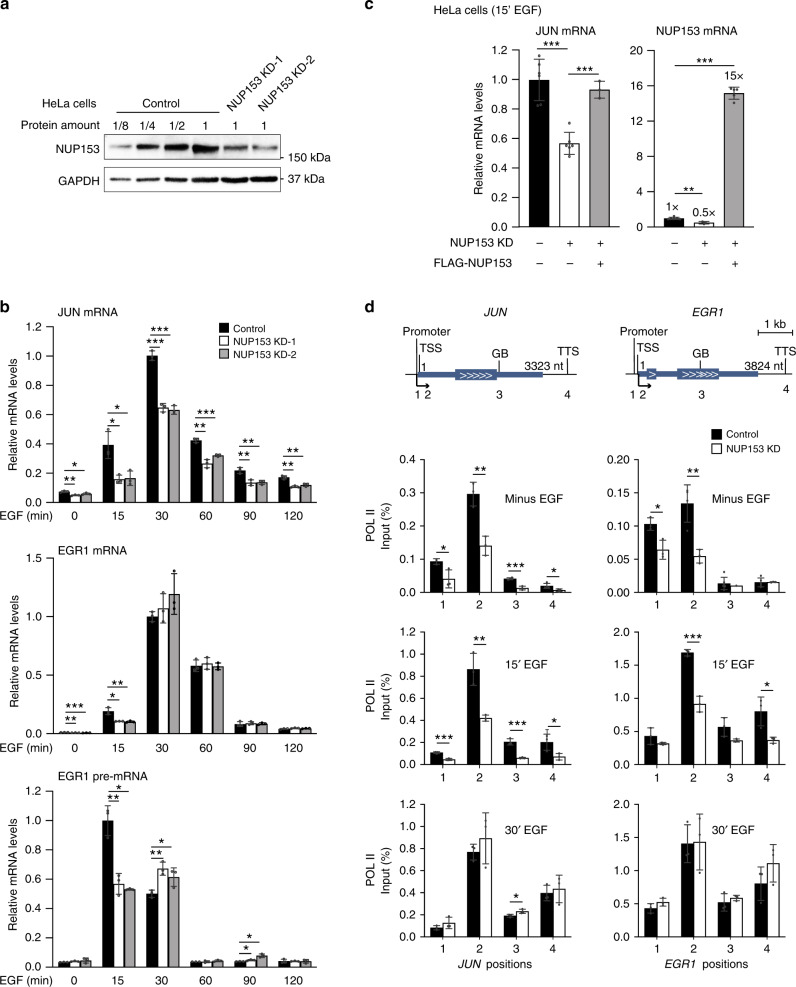
Fig. 4. NUP153 controls POL II recruitment to the IEG promoters and impacts IEG transcription initiation.
a Western blot showing NUP153 protein levels in control and NUP153 KD HeLa cells. b Real-time RT-PCR showing relative IEG mRNA and nascent mRNA levels in control and NUP153 KD HeLa cells in a time course dependent manner. JUN mRNA levels at minus EGF (control vs KD-1, **p = 0.0033; control vs KD-2, *p = 0.0345), 15 min EGF (control vs KD-1, *p = 0.0138; control vs KD-2, *p = 0.0199), 30 min EGF (control vs KD-1, ***p = 0.0001; control vs KD-2, ***p = 0.0001), 60 min EGF (control vs KD-1, **p = 0.0010; control vs KD-2, ***p = 0.0004), 90 min EGF (control vs KD-1, **p = 0.0065; control vs KD-2, **p = 0.0045), 120 min EGF (control vs KD-1, **p = 0.0010; control vs KD-2, **p = 0.0050). EGR1 mRNA levels at minus EGF (control vs KD-1, **p = 0.0018; control vs KD-2, ***p = 0.0008), 15 min EGF (control vs KD-1, *p = 0.0373; control vs KD-2, **p = 0.0066). EGR1 pre-mRNA levels at 15 min EGF (control vs KD-1, **p = 0.0038; control vs KD-2, *p = 0.0151), 30 min EGF (control vs KD-1, **p = 0.0045; control vs KD-2, *p = 0.0436), 90 min EGF (control vs KD-1, *p = 0.0364; control vs KD-2, *p = 0.0305). c Real-time RT-PCR showing relative JUN and NUP153 mRNA levels in control, NUP153 KD and FLAG-NUP153 expressing NUP153 KD HeLa cells upon 15 min EGF treatment. GAPDH was used to normalize mRNA levels. JUN mRNA levels in control vs KD (***p = 0.0000); KD vs FLAG-NUP153 (***p = 0.0001). NUP153 mRNA levels in control vs KD (**p = 0.0011); KD vs FLAG-NUP153 (***p = 0.0000). d POL II binding across JUN and EGR1 genetic elements was mapped by ChIP real-time PCR in control and NUP153 KD HeLa cells under indicated conditions (see Supplementary Data 10 for primer sequences). POL II binding at JUN, minus EGF (control vs KD; promoter, *p = 0.0329; TSS, **p = 0.0042; GB, ***p = 0.0003; TTS, *p = 0.0493), 15 min EGF (control vs KD; promoter, ***p = 0.0009; TSS, **p = 0.0065; GB, ***p = 0.0009; TTS, *p = 0.0443), 30 min EGF (control vs KD; GB, *p = 0.0385). POL II binding at EGR1, minus EGF (control vs KD; promoter, *p = 0.0172; TSS, **p = 0.0060), 15 min EGF (control vs KD; TSS, ***p = 0.0004; TTS, *p = 0.0281). Data shown are percent (%) of input. Values are mean ± standard deviation. Two-tailed Student’s t-test, n ≥ 3 independent experiments. Nt, nucleotide. Source data are provided as a Source Data file.