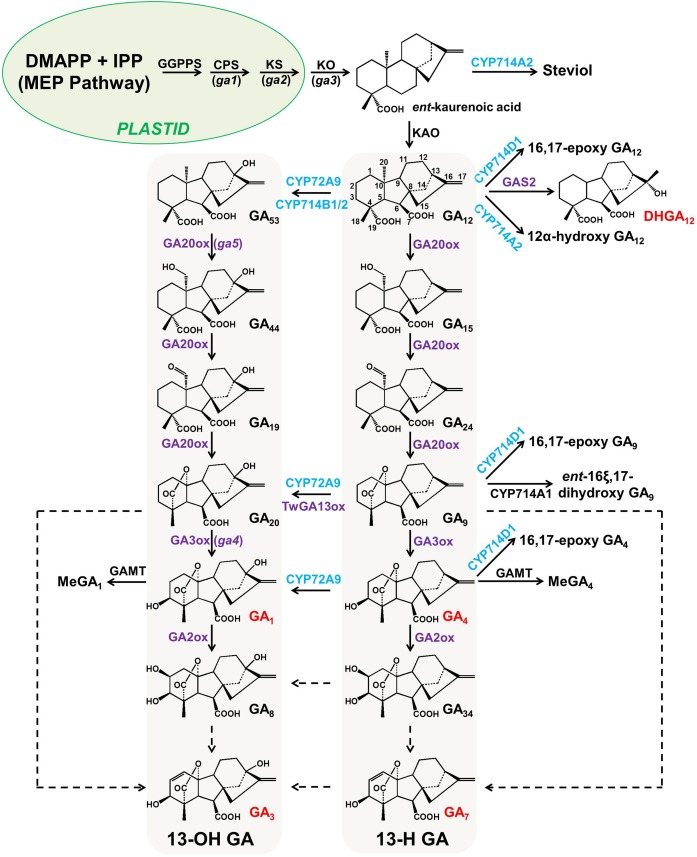
FIGURE 1.
Simplified GA metabolic pathway in flowering plants. All enzymes mapped to the GA metabolic pathway, except GA13ox from Tripterygium wilfordii, were verified with enzymatic assays and the chemical profiling of transgenic plants. P450s (ER membrane-associated proteins) in this pathway are highlighted in blue, while 2-oxoglutarate-dependent dioxygenases (2-ODDs, cytosolic proteins) are highlighted in purple. All dashed lines indicate enzymatic a step that has not yet been characterized at the genetic level in any flowering plant. The genes/enzymes characterized from Arabidopsis and rice are marked in black and blue, respectively. The carbon backbone of GA12 is labeled with numbers, and the bioactive GAs (GA1, GA3, GA4, GA7, and DHGA12) are marked in red. The Arabidopsis GA-deficient mutants (ga1 to ga5) used for map-based cloning are shown in parentheses next to the corresponding genes. CPS, ent-copalyl diphosphate synthase; GAMT, GA methyltransferase; GAS2, gain of function in ABA-modulated seed germination 2; GGPPS, geranylgeranyl diphosphate synthase; KAO, ent-kaurenoic acid oxidase; KO, ent-kaurene oxidase; KS, ent-kaurenoic acid synthase; MEP, 2-C-methyl-D-erythritol 4-phosphate.