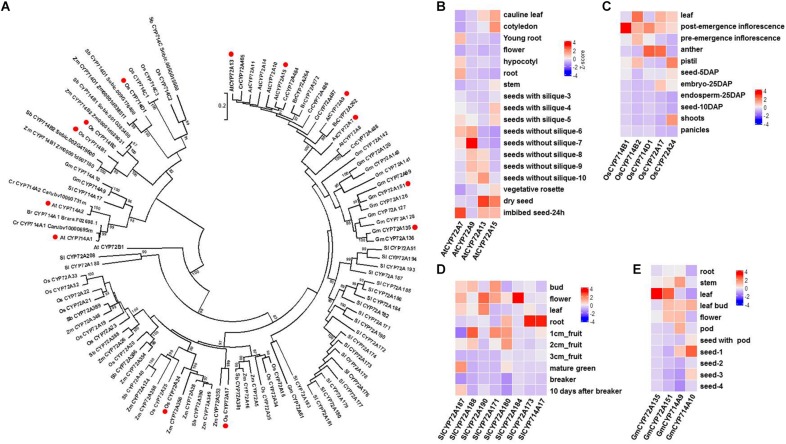
FIGURE 2.
Characterization of GA oxidases encoded by P450 genes in flowering plants. (A). Phylogenetic analysis of plant GA oxidases encoded by P450 genes using MEGA 6.0 (Tamura et al., 2013). A total of 102 P450 proteins (members of the CYP72A and CYP714 subfamilies) were obtained from 8 representative species in plant evolutionary history. P450 proteins with GA oxidase activity are marked with red circles. Bootstrap values (based on 500 replicates) >80% are shown for corresponding nodes. At, Arabidopsis thaliana; Br, Brassica rapa; Cr, Capsella rubella; Gm, Glycine max; Os, Oryza sativa; Sb, Sorghum bicolor; Sl, Solanum lycopersicum; Zm, Zea mays. (B) Tissue specificity of four CYP72A genes in Arabidopsis. Data were extracted from the gene expression map of Arabidopsis development (http://arabidopsis.org). (C) Tissue specificity of three CYP714 genes and two CYP72A genes in rice. RNA-Seq (FPKM) data were extracted from the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu). (D) Tissue specificity of seven CYP72A genes and one CYP714A gene in tomato plants. Data were extracted from the Tomato Genome Consortium (Sato et al., 2012). (E) Tissue specificity of two CYP72A genes and two CYP714A genes in soybean plants. Data were extracted from Shen et al. (Shen et al., 2014).