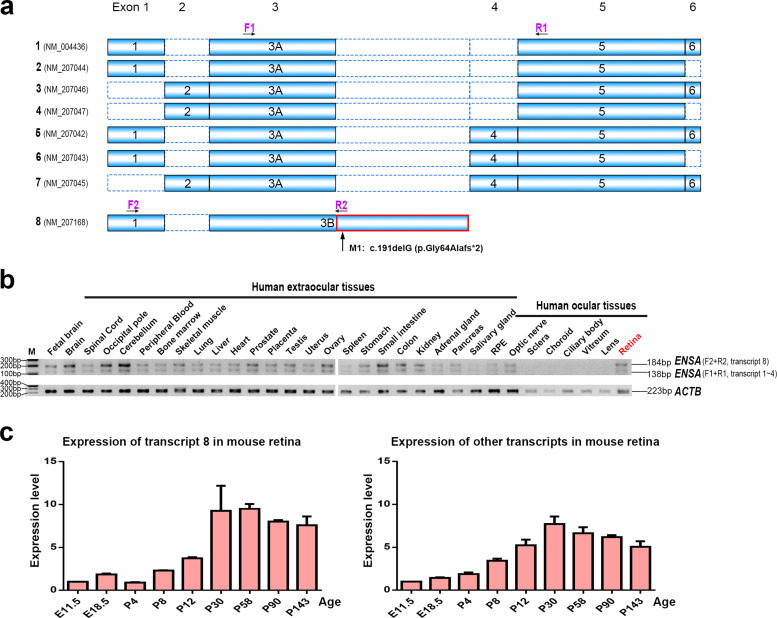
Fig. 2.
Transcripts and expression analysis of ENSA mRNA. (a) The coding scheme of eight alternative transcripts of ENSA. Transcript 8 (NM_207168) is composed of exons 1 and exon 3B. The mutation in ENSA, c.191delG (p.Gly64Alafs*2), is located at exon 3B of transcript 8 (the intronic +8 position of the alternative exon 3A in other transcripts). The differences between exon 3B and exon 3A are indicated in red (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article). Arrows represent the location of primers used for RT-PCR (see also Supplementary Materials, Table S2). (b) The RT-PCR of ENSA in 32 human tissues. Two sets of primers were used to amplify different transcripts of ENSA: set 1 (F1+R1), representing transcripts 1~7, and set 2 (F2+R2), representing transcript 8. ACTB amplification served as a control for cDNA amount in the PCR analysis. It should be noted that transcripts 5, 6, and 7, in which an additional 48 bp exon (exon 4) is encompassed, have been deposited in GenBank, but that we were not able to verify it in any of the analyzed tissues. M, marker. (c) Expression of Ensa in mouse retinas at different developmental stages by qPCR. Ex, embryonic x days. Px, x days after birth. Each of the RNA samples was obtained from three wild-type mice (six retinas). The experiments were repeated 3 times. β-Actin was used for normalization. Error bars represent the standard errors of the relative quantities.