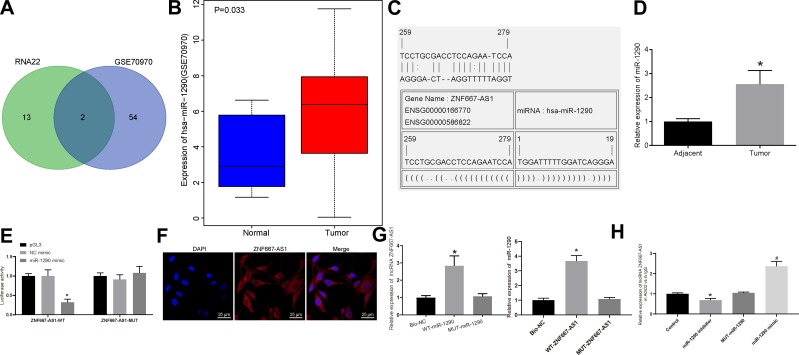
Figure 3.
ZNF667-AS1 competitively binds to miR-1290. (A) Comparison of miRNA results with the possible binding of ZNF667-AS1 predicted by RNA22 and miRNAs up-regulated in GSE70970 microarray dataset, there are 2 miRNAs in the intersection. (B) The expression of miR-1290 in GSE70970 microarray dataset. (C) RNA22 predicts the binding site between ZNF667-AS1 and miR-1290. (D) RT-qPCR utilized to detect the expression of miR-1290 in tumor and adjacent tissues (n = 36), *p < 0.05 vs adjacent tissues. (E) Dual-luciferase reporter gene assay to verify the binding relationship between ZNF667-AS1 and miR-1290. (F) FISH for the detection of subcellular localization of ZNF667-AS1 in CNE-1 cells (×200). (G) RNA pull-down assay to verify the binding of ZNF667-AS1 with miR-1290, *p < 0.05 vs the Bio-NC group. (H) RIP assay for the detection of whether ZNF667-AS1 interacted directly with AGO2 protein in CNE-1 cells, *p < 0.05 vs the IgG group; #p < 0.05 vs the MUT-miR-1290 group. Data are expressed as mean ± standard deviation. Paired t test is used for comparison of two groups. One-way analysis of variance is used for comparison among multiple groups. Each experiment was run in triplicate.
Abbreviations: ZNF667-AS1, zinc finger protein 667-antisense RNA 1; miR-1290, microRNA-1290; RT-qPCR, reverse transcription quantitative polymerase chain reaction; FISH, fluorescence in situ hybridization; NC, negative control; RIP, RNA immunoprecipitation; AGO2, argonaute 2; IgG, immunoglobulin G; WT, wild type; MUT, mutant; DAPI, 4ʹ,6-diamidino-2-phenylindole.