Abstract
The North European species of Elaphomyces section Elaphomyces (Eurotiales, Pezizomycotina) are studied. Three new species, E. citrinopapillatus, E. pusillus, and E. roseoviolaceus are introduced and verified by morphology and sequence data from ITS, nuclear LSU, mitochondrial SSU, and β-tubulin. A lectotype for Elaphomyces granulatus is selected. Elaphomyces granulatus and E. muricatus are epitypified with sequenced material from the Femsjö region in South Sweden. Elaphomyces striatosporus is epitypified with sequenced material from the vicinity of the type locality in Norway. A key to all species of Elaphomyces occurring in Denmark, Norway, and Sweden is provided.
Keywords: Eurotiaceae, Eurotiomycetes, key, multi-gene phylogeny, nomenclature, Pezizomycotina, three new taxa
INTRODUCTION
The genus Elaphomyces (Nees von Esenbeck & Nees von Esenbeck 1820) comprises hypogeous species forming ectomycorrhiza with a variety of forest trees. The genus was described based on a single species, Scleroderma cervinum, ascribed to Persoon. The genus name was later sanctioned by Fries (1829), who listed two species, E. granulatus (= Scleroderma cervinum) and E. muricatus. Index Fungorum (2018) counts nearly 100 described names but recent studies have estimated the number of valid species to be around 55 on a global scale (Castellano et al. 2012a). Early mycologists working on the European species of Elaphomyces were Vittadini (1831, 1842) and Tulasne & Tulasne (1841, 1851) and in their works several of the common and widespread species were described. In a comprehensive study of the genus Ławrynowicz (1988, 1989) recognized 20 species occurring in Europe. More recently Paz et al. (2017) combined morphology and molecular data in a revision of the European taxa and accepted 26 species. The genus has a global distribution and is recorded from all continents, except the Antarctic (Zhang & Minter 1989, Zhang 1991, Castellano et al. 2011, 2012a–c, 2016, Buyck et al. 2016, Castellano & Stephens 2017). Molecular phylogenetic studies indicate that Elaphomyces, as it is presently understood, covers a considerable genetic variation and its monophyly has been questioned (Reynolds 2011, Buyck et al. 2016, Paz et al. 2017).
Vittadini (1831) divided Elaphomyces in two major sections, Malacodermei and Sclerodermei. Within the latter section he distinguished two species groups: species with a more or less smooth peridium (Cortice laevi) and those with a verrucose peridium (Cortice exasperato). Fontana (1909), when dealing with Vittadini’s group “cortice exasperato”, distinguished two lineages: species with a non-homogeneous peridium (the E. variegatus-group = E. muricatus and its allies) and those with a homogenous peridium (the E. granulatus-group). Phylogenetic analyses based on molecular data show that Fontana’s two groups both belong to a well-supported clade that corresponds to Elaphomyces sensu stricto (= section Elaphomyces) (Buyck et al. 2016, Castellano et al. 2016, Paz et al. 2017). This section covers species with a mostly more or less brown peridial surface. In accordance with Fontana (1909) the molecular data confirm a division of sect. Elaphomyces in two subsections, Muricati and Elaphomyces, respectively. In addition, a third subsection, sect. Papillati has been recognized (Paz et al. 2017).
The present study extends on the work presented in Paz et al. (2017) and focuses on section Elaphomyces in North Europe (Denmark, Norway, Sweden). Truffle inventories using trained dogs have been undertaken in a variety of ecosystems. From newly collected ascomata a four-gene (ITS, nLSU, mtSSU and -tubulin) sequence data set was generated and analysed by phylogenetic methods. The aims were to survey the diversity of Elaphomyces in the countries mentioned, relate all specimen sequences to existing names, describe new species when necessary, and infer phylogenetic relationships among the species.
MATERIALS AND METHODS
Specimen sampling
Our sampling has an emphasis on Elaphomyces in Europe and its northern part in particular, with the aim of finding and characterising all known or putative species of Elaphomyces subgenus Elaphomyces occurring in the Nordic countries. Representatives of Pseudotulostoma and Aspergillus were selected as outgroup taxa in the analyses. Sequence data of these were retrieved from GenBank and added to the dataset (FJ358278, AB161194). Two ITS sequences of E. muricatus and E. granulatus were retrieved from GenBank and included in the dataset (EU784198, EU784197). In addition, sequence data from southern Europe and type specimens in the study by Paz et al. (2017) were included for comparison of species concepts and genetic variation (Table 1). For the epitypification of species originally described by Fries from Sweden, new specimens from Femsjö, Småland, the place where Fries was collecting, were sampled and sequenced.
Table 1.
Specimens of Elaphomyces spp. used for phylogenetic analyses, and their corresponding GenBank accession numbers.
| Species | Status | Voucher ID | Country, year | Herbarium | ITS | LSU | mtSSU | β-tubulin |
|---|---|---|---|---|---|---|---|---|
| Elaphomyces asperulus | Epitype | IC13051208 | Spain, 2012 | LIP-0001131 | KX238833 | KX238877 | ||
| A. Molia, AM-35-2014 | Sweden | GB-0150464 | KR029753 | KR029753 | KR064772 | KR363209 | ||
| A. Molia s.n. | Denmark, 2014 | O-F22178 | KR029755 | KR029755 | KR064773 | KR363210 | ||
| A. Molia, AM-179-2013 | Norway, 2013 | O-F21354 | KR029754 | KR029754 | – | – | ||
| G.F. Medardi | Italy | MCVE-00160 | KR064762 | KR064762 | – | – | ||
| Elaphomyces barrioi | Holotype | IC16121209 | Spain, 2012 | LIP-0001133 | KX238848 | – | – | – |
| A. Molia, AM-270a-2014 | Norway, 2014 | O-F22181 | KR029744 | KR029744 | KR064767 | – | ||
| A. Molia, AM-184-2014 | Norway, 2014 | O-F22180 | KR029746 | KR029746 | KR064768 | – | ||
| A. Molia, AM-347-2011 | Norway, 2011 | O-F21187 | KR029745 | KR029745 | – | – | ||
| A. Molia, AM-32-2014 | Norway, 2014 | O-F22301 | KR029747 | KR029747 | KR064769 | KR363206 | ||
| Elaphomyces cf. barrioi | A. Molia, AM 153 | Norway, 2011 | A Molia pers. herb. | KR076543 | – | – | – | |
| Elaphomyces citrinopapillatus | Holotype | A. Molia, AM-23-2014 | Norway, 2014 | O-F21559 | KR029765 | KR029765 | – | – |
| A. Molia, AM-69-2014 | Norway, 2014 | O-F22184 | KR029762 | KR029762 | KR064778 | KR363213 | ||
| A. Molia, AM121a-2013 | Norway, 2013 | O-F21344 | KR029763 | KR029763 | KR064779 | KR363214 | ||
| A. Molia, AM-19-2014 | Norway, 2014 | O-F21561 | KR029766 | KR029766 | – | – | ||
| A. Molia, s.n. | Norway, 2013 | O-F21556 | KR029764 | KR029764 | – | – | ||
| Elaphomyces decipiens | Neotype | IC28011203 | Spain, 2012 | LIP-0001134 | KX238832 | KX238876 | – | – |
| IC27111118 | Spain, 2011 | A. Paz pers. herb. | KX238842 | – | – | – | ||
| Elaphomyces cf. decipiens | IC12051208 | Spain, 2012 | A. Paz pers. herb. | KX238831 | – | – | – | |
| A. Molia et at. s.n. | Norway, 2013 | O-F21513 | KR029743 | KR029743 | KR064766 | KR363205 | ||
| M. Jeppson, MJ10151 | Sweden | GB | MF614923 | – | – | – | ||
| A. Molia, AM-351-2013 | Norway, 2013 | O-F21484 | KR029742 | KR029742 | KR064765 | KR363204 | ||
| Elaphomyces granulatus | Epitype | A. Molia, AM-44-2014 | Sweden, 2014 | GB-0147063 | KR029767 | KR029767 | – | – |
| M.C. Clark | Scotland, 1972 | K(M)47712 | EU784197 | – | – | – | ||
| A. Molia, AM-20-2012 | Norway, 2012 | O-F245217 | KR029768 | KR029768 | – | KR363215 | ||
| Elaphomyces granulatus f. pallidosporus | Holotype | IC21071103 | Italy, 2011 | LIP-0001132 | KX238846 | – | – | – |
| Elaphomyces muricatus | Epitype | A. Molia, AM-43-2014 | Sweden, 2014 | GB-0147062 | KR029730 | KR029730 | – | KR363200 |
| A. Molia, AM-37-2014 | Sweden, 2014 | GB | KR029731 | KR029731 | – | KR363201 | ||
| A. Molia, AM-42-2014 | Sweden, 2014 | GB | KR029732 | KR029732 | KR064763 | KR363202 | ||
| A. Molia, AM-157-2012 | Norway, 2012 | O-F245291 | KR029733 | – | – | |||
| A. Molia, AM-264-2014 | Norway, 2014 | O-F22182 | KR029734 | KR029734 | – | – | ||
| A. Molia, s.n. | Norway, 2014 | O-F22183 | KR029735 | KR029735 | – | – | ||
| IC01041301 | Spain, 2013 | A. Paz pers. herb. | KX238849 | – | – | – | ||
| M. Kelly | England, 2004 | K(M)121442 | EU784198 | |||||
| A. Molia, AM-212-2013 | Norway, 2013 | O-F21312 | KR029741 | KR029741 | KR064764 | KR363203 | ||
| Elaphomyces muricatus var. reticulatus | Epitype | IC14011206 | Spain, 2012 | LIP-0001153 | KX238851 | – | – | – |
| Elaphomyces muricatus var. variegatus | Epitype | IC05011307 | Spain, 2013 | LIP-0001154 | KX238850 | – | – | – |
| A. Molia, AM-351-2011 | Norway, 2011 | O-F21190 | KR029737 | KR029737 | – | – | ||
| A. Molia, AM-239-2012 | Norway, 2012 | O-F245312 | KR029736 | – | – | – | ||
| Elaphomyces cf. muricatus | R. Kristiansen s.n. | Norway, 2011 | O-F245437 | KR029738 | – | – | – | |
| A. Molia, AM-352-2011 | Norway, 2011 | O-F21009 | KR029739 | KR029739 | – | – | ||
| A. Molia, AM-151-2014 | Sweden, 2014 | GB | KR029740 | KR029740 | – | – | ||
| Elaphomyces papillatus var. papillatus | Epitype | IC12051202 | Spain, 2012 | LIP-0001136 | KX238819 | KX238872 | – | – |
| IC26051201 | Spain, 2012 | A. Paz pers. herb. | KX238820 | – | – | – | ||
| Elaphomyces papillatus var. suphureopallidus | Holotype | IC13051212 | Spain, 2012 | LIP-0001156 | KX238830 | – | – | – |
| Elaphomyces pusillus | Holotype | A. Molia, AM 121 | Norway, 2014 | O-F22174 | KR029761 | KR029761 | KR064777 | KR363212 |
| A. Molia, AM 132-2014 | Sweden, 2014 | GB-0179907 | KR029758 | KR029758 | KR064774 | – | ||
| A. Molia, AM-123-2014 | Norway, 2014 | O-F22175 | KR029760 | KR029760 | KR064776 | KR363211 | ||
| S. Sivertsen, B.K.P. Sveum 79-205 | Norway, 1979 | TRH-1273 | KR029759 | – | KR064775 | – | ||
| Elaphomyces cf. pusillus | A. Molia, AM-146-2012 | Norway, 2012 | O-F245285 | KR029756 | KR029756 | – | – | |
| K. Killingmo, AKW-421 | Norway, 2012 | O-F21005 | KR029757 | KR029757 | – | – | ||
| Elaphomyces roseoviolaceus | Holotype | A. Molia, AM-135-2013 | Norway, 2013 | O-F21376 | KR029752 | KR029752 | KR064770 | KR363207 |
| A. Molia, AM-271-2013 | Norway, 2013 | O-F21429 | KR029751 | KR029751 | KR064771 | KR363208 | ||
| A. Molia, AM-271-2013 | Norway, 2013 | O-F21429 | KR029750 | KR029750 | – | – | ||
| Elaphomyces quercicola | Holotype | IC23071107 | Spain, 2011 | LIP-0001155 | KX238837 | KX238879 | – | – |
| IC23071104 | Spain, 2011 | A. Paz pers. herb. | KX238838 | – | – | – | ||
| Elaphomyces striatosporus | Epitype | A. Molia, AM-269-2012 | Norway, 2012 | O-F245330 | KR029748 | – | – | – |
| A. Molia, AM-273-2012 | Norway, 2012 | O-F245333 | KX238861 | – | – | – | ||
| A. Molia, AM-280-2012 | Norway, 2012 | O-F245337 | KR029749 | – | – | – | ||
| Elaphomyces violaceoniger | Holotype | IC22011401 | Spain, 2011 | LIP-0001135 | KX238857 | – | – | – |
| IC15031401 | Norway, 2014 | A. Paz pers. herb. | KX238858 | – | – | – |
DNA extraction, amplification (PCR), and sequencing
Sequences from four regions were generated: the complete ITS region and about 1 200 base pairs (bp) of the 5ʹend of the LSU nuclear ribosomal DNA, the mitochondrial small subunit ribosomal DNA (mtSSU) and about 500 bp of the β-tubulin (β-tub) gene. DNA extractions, PCR reactions, and sequencing were performed as described in Larsson & Örstadius (2008). Primers used to amplify the complete ITS region and the 5ʹend of the LSU region were ITS1F (Gardes & Bruns 1993) and LR21, LR0R, and LR7 (Hopple & Vilgalys 1999); for mtSSU we used MS1 and MS2 (White et al. 1990); for β-tub Bt1a and Bt2b (Glass & Donaldson 1995). Primers used for sequencing were ITS1, ITS4, MS1, MS2 (White et al. 1990), Ctb6 (https://nature.berkeley.edu/brunslab/) and Lr5 (Hopple & Vilgalys, 1999), Bt2a and Bt2b. Some of the DNA barcode data were generated in collaboration with the Norwegian Barcode of Life project (NorBOL). Sequenced specimens are marked with an asterisk (*) in specimen lists.
Phylogenetic analyses
Sequences were edited and assembled using Sequencher v. 5.1 (Gene Codes, Ann Arbor). Alignment of individual genes was performed using the L-INS-i strategy as implemented in MAFFT v. 7.017 (Katoh & Standley 2013). The alignment was adjusted manually using the data editor in PAUP v. 4.0b10 (Swofford 2003). The sequences generated for this study have been deposited in GenBank with the following accession numbers: ITS-LSU KR029730-KR029767 and MF614923, mtSSU KR064762-KR064785, β-tub KR363193-363215 (Table 1). Some of these sequences were also used in the analyses carried out by Paz et al. (2017).
Sequences were concatenated and subjected to phylogenetic analyses through maximum parsimony and Bayesian inference. Variable regions with ambiguous alignment were excluded and gaps were treated as missing data. Heuristic searches for the most parsimonious trees were performed using PAUP (Swofford 2003). All transformations were considered unordered and equally weighted. Heuristic searches with 1 000 random-addition sequence replicates and TBR branch swapping were performed, saving at most 25 trees in each replicate. Relative robustness of clades was assessed by the bootstrap method using 1 000 heuristic search replicates with 100 random taxon addition sequence replicates and TBR branch swapping, the latter saving at most 25 trees in each replicate.
Bayesian analysis was carried out in MrBayes v. 3.2.6 (Ronquist et al. 2012), with a best-fit model of nucleotide evolution supplied separately for each genetic marker by MrModeltest v. 2.2 (Nylander 2004). The protein-coding β-tubulin gene was not subjected to partitioning of the third base in each codon. Eight default-setting Metropolis-Coupled Markov Chain Monte Carlo (MCMCMC) chains were run for 10 M generations with trees sampled every 5 000 generations and an initial burn-in of 1 000 trees. After discarding the trees prior to the burn-in threshold a 50 % majority-rule consensus phylogram was computed from the remaining trees.
Morphology
Ascomata of Elaphomyces were detected in the field, typically with the help of trained dogs. The samples were photographed in situ and later studied in the laboratory. The features of the peridial surface were studied after the ascomata had been carefully cleansed from soil, using a tooth brush. Ascomata were measured from fresh material, using a caliper. The measurements of the peridial thickness refer to fresh material and always include the outermost peridial layer (cortex). If fresh material was not available, dried samples were soaked in water for at least 30 minutes before measuring. Spore measurements are given inclusive of their ornamentation and were made at 1 000×, using a Leica DM LS microscope. Twenty mature (or putatively mature) spores with well-developed ornamentation were measured for each sample. All microscopic studies were conducted in Hoyer’s medium (http://www.coloss.org/beebook/II/varroa/2/3/1/2). Micrographs were taken in a Zeiss AXIO Imager M2, using the software Zeiss ZEN 2 pro. Collections have been deposited at herbarium O, if not otherwise stated.
RESULTS
Phylogenetic analyses
The complete concatenated and aligned four-gene dataset consisted of 63 taxa and 3 165 nucleotide positions. After exclusion of ambiguous regions 2 871 characters remained for the analyses. Of these 2 510 were constant, 99 were variable and parsimony uninformative, and 262 were parsimony informative. The Maximum parsimony analysis yielded 23 225 equally most parsimonious trees (length = 479, CI = 0.8601, RI = 0.9712) one of which is presented in Fig. 1. Bootstrap values above 50 % are indicated above branches. A bootstrap value greater than 70 % is considered strong.
Fig. 1.
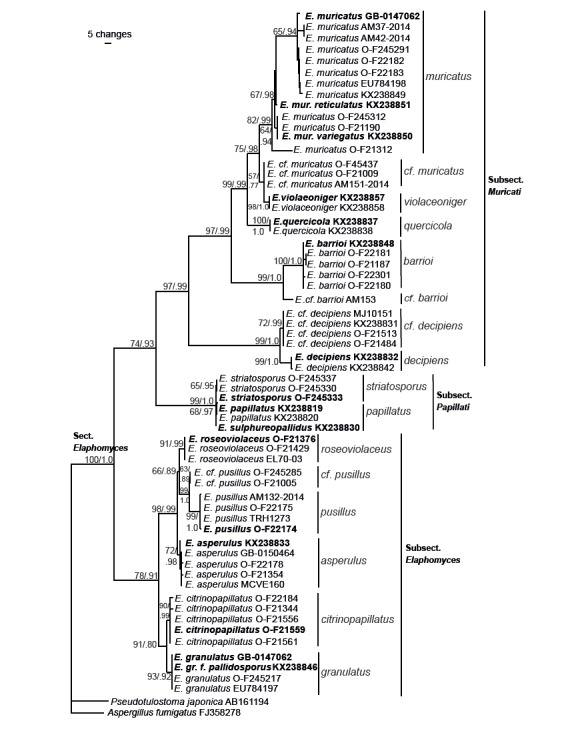
One of the equally most parsimonious trees from the phylogenetic analysis based on nuclear rDNA ITS and partial LSU, mtSSU, and β-tubulin sequences. Parsimony bootstrap values and Bayesian posterior probabilities are indicated on branches. Clades discussed in the text are indicated with bars and species epithets. Sequences originating from type specimens are marked in bold.
As suggested by MrModeltest (Nylander 2004), the following nucleotide evolution models were used in the partitioned Bayesian analysis: GTR+I for ITS1, JC+I for 5.8S, HKY+G for ITS2, GTR+I+G for LSU, GTR+I for mtSSU, and K80+G for β-tubulin. The MCMC analysis converged well in advance of the burn-in threshold and chain mixing was found to be satisfactory, as assessed by Tracer v. 1.5 (Drummond et al. 2012). Also in the Bayesian analysis, section Elaphomyces was recovered as monophyletic with strong support (BPP 1.00). The Bayesian tree topology is identical to the maximum parsimony tree. The same subclades and terminal clades supported in the bootstrap analysis were also recovered and supported in the Bayesian analysis. BPP values are indicated on the corresponding branches in Fig. 1. A BPP value above 0.95 is considered significant.
The analyses recovered section Elaphomyces (100/1.0) as monophyletic and divided in three supported subclades corresponding to subsection Elaphomyces (78/0.91), subsection Papillati (99/1.0), and subsection Muricati (97/0.99). Within section Elaphomyces 15 terminal clades and one single branch, weakly to strongly supported, were recovered. These correspond to E. muricatus (82/0.99), E. cf. muricatus (57/.77), E. violaceoniger (98/1.0), E. quercicola (100/1.0), E. barrioi (100/1.0), E. cf. barrioi (99/1.0), E. decipiens (99/1.0), E. cf. decipiens (72/0.99), E. papillatus (68/0.97), E. striatosporus (65/0.95), E. roseoviolaceus (91/0.99), E. pusillus (99/1.0), E. cf. pusillus (63/0.89), E. asperulus (72/0.98), E. citrinopapillatus (90/0.99), and E. granulatus (93/0.92) (Fig. 1).
The terminal clades corresponding to new species are described and discussed in the Taxonomy section. Also, typification measures are presented here. An overview of all species in section Elaphomyces is given in the Discussion section. For morphological descriptions of already described species in section Elaphomyces we refer to Paz et al. (2017).
Taxonomy
Elaphomyces citrinopapillatus A. Molia, A. Paz & Lavoise, sp. nov. Figs 2–3. MycoBank MB833573.
Fig. 2.

Elaphomyces citrinopapillatus. Holotype (O-F21559). A. Mature ascomata. B. Section of peridium. C. Detail of cortex. D. Peridial structure. E. Section of cortex. F, G. Ascospores. Scale bars: A, C = 10 mm; B = 1 mm; D–G = 20 µm.
Fig. 3.

Type locality of Elaphomyces citrinopapillatus. Norway, Akershus, Nittedal, Slattum, 10 Jan. 2014.
Etymology: citrinus (lemon yellow) and papillatus (warty), referring to the more or less enclosed, yellow warts of the peridium.
Typus: Norway, Akershus, Nittedal, Slattum (WGS84: N60.00808 E10.91350), in moist old-growth Picea stand on rich calcareous soil with moss cover, 10 Jan. 2014, Lello (dog) & A. Molia, holotype in herb. O, O-F21559*, barcode sequence GenBank KR029765.
Ascomata globose to depressed globose, 0.6–2.0(–2.6) cm (av. 1.3 cm, n = 310), black when fresh, dull brownish black when dry, with a brown, thin and ephemeral hyphal envelope. Odour strong and unpleasant, metallic. Peridium in section (0.65–)0.7–1.4 mm thick, white to cream, towards the gleba "café au lait"-colour, with enclosed, lemon yellow, rounded to elongated or ± conical warts, about 250 µm high, breaking through the cortex. After cleansing of the ascomata these warts are easily detected on the brownish black peridial surface. Cortex 350–500 µm thick, sharply delimited from the underlying peridial layer. The inner part of the peridium is constructed of loosely interwoven, thick-walled (up to 2 µm), angularly rounded to elongated cells. Near the gleba these cells are brownish and sometimes confluent and more or less shapeless. The exterior peridial layer, the cortex, is blackish brown, constructed of rounded and thick-walled cells on the surface, turning pyramidal to elongated inwards. Gleba black upon maturity, pulverulent. Young and immature ascomata have a continuous fleshy texture with coloured hyphae surrounding lumps of developing spore mass (Fig. 2). Asci 8-spored. Ascospores globose, (21–)23–32(–33) µm (av. 26 µm, n=240), brown, echinulate, spines 1.5–2.5 µm high, well separated and not coalescing.
Ecology and distribution: Develops gregariously in the upper soil layer (black to dark brown soil) in semi-rich to rich woodlands. It occurs in pure Picea abies stands, but also in mixed forests with Picea abies, Pinus sylvestris, Betula spp. and Corylus avellana. It is to date known from five counties in southern Norway. All sites were located by trained dogs.
Additional materials examined: Norway, Akershus, Nittedal, Slattum, in old, rich Picea abies forest, 10 Jan. 2014, Lello (dog) & A. Molia, O-F21560, O-F21561*, O-F21392; Akershus, Oppegård, Svartskog, by Svartskog kindergarten, in semi-rich mixed forest, under Picea abies, 9 Aug. 2014, Lello (dog), P.-A. Moreau & A. Molia, O-F22129; Buskerud, Flesberg, Lyngdal, Molia, Tritjenna, in semi-rich Picea abies forest with Betula, near forest road, 27 Dec. 2013, Lello (dog) & A. Molia, O-F21556*; ibid. in semi-rich Picea abies forest, 17 Apr. 2014, Lello (dog) & A. Molia, O-F22184*; ibid., in young, semi-rich Picea abies forest near forest road, A. Molia 26–2015 (GB); ibid., Botnan, in rich Picea abies forest, 3 Nov. 2013, Lello (dog) & A. Molia, O-F21458; Flesberg, Sølset, towards Hommelisetera, in semi-rich forest with Picea abies, Salix sp. and Betula sp., 18 Apr. 2016, Lello (dog) & A. Molia, O-F22515; Sigdal, Hagavollesetra, in semi-rich Picea abies forest, 2 Nov. 2013, Lello (dog), Å. Borge & A. Molia, O-F21457; Oslo, Sognsvann, in semi-rich Picea abies forest, 13 Sep. 2013, Lello (dog) & A. Molia, O-F21303, F21304; ibid., in semi-rich Picea abies forest, 25 Aug. 2013, Lello (dog), T. Læssøe & A. Molia, O-F21344*; Oslo, Ekebergsåsen, in semi-rich conifer forest under Picea abies, 12 Jul. 2012, Kokkos & Viktoria (dogs), K. Killingmo, M. Jeppson & A. Molia, O-F245249, F245254; ibid. 18 Jul. 2012, O-F245258, O-F245258; Østfold, Marker, S of Ørje, in semi-rich Picea abies forest, 13 Apr. 2014, Louise (dog), K. Killingmo & A. Molia, O-F22128; Telemark, Bamble, Røsskleiva, rich mixed forest, under Picea abies, 10 Oct. 2011, Lello (dog), A.K. Wollan & A. Molia, (O-F21189). ibid., 10 Oct. 2011, Kokkos, Viktoria (dogs), K. Killingmo & T. Andersen (O-F21194); Skien, Luksefjell-Ulfskollen, semi-rich forest with Populus tremula, Corylus avellana, Quercus sp. and Picea abies, 27 Sep. 2015, Lello (dog), A. Molia & B.F. Høifødt, (O-F260438); Porsgrunn, Frierflauane, rich mixed forest, under Picea abies, 28 Sep. 2013, A. Paz, O-F21277; ibid., Frierstien, in mixed rich forest, under Picea abies, 2 Oct. 2015, Lello (dog) & A. Molia, O-F260439; Tinn Austbygd, Mæl, in rich mixed forest, under Picea abies, 18 Jun. 2015, Lello (dog) & A. Molia, O-F22483.
Remarks: Elaphomyces citrinopapillatus is usually easy to recognize already in the field due to its unique features of the peridium. The closely related species E. asperulus, E. granulatus, E. pusillus and E. roseoviolaceus also have yellowish peridial warts but with less contrasting colours. A detailed study of the literature did not reveal a suitable name for this species. Our molecular data confirm it as a distinct species, sister to E. granulatus, from which it differs by generally smaller ascomata, the strongly contrasting yellow warts and the production of gregarious ascomata.
Elaphomyces granulatus Fr., Systema mycologicum (Lundae) 3: 58. 1829. Figs 4, 9A–B.
Fig. 4.

Elaphomyces granulatus. Lectotype, third basidioma from left (UPS, F-708238).
Fig. 5.

Elaphomyces pusillus. Holotype (O-F22174). A. Mature ascomata B. Detail of cortex. C. Ascocarp in section. D. Peridial structure. E. Section of peridium showing peridial warts (yellowish). F. Peridial structure between warts. G, H. Ascospores. Scale bars: A–C = 10 mm; D = 10 µm; E–H = 20 µm.
Fig. 6.

Type locality of Elaphomyces pusillus. Norway, Nord-Trøndelag, Verdalen, Inndalen, 8 Sep. 2014.
Fig. 7.

Elaphomyces roseoviolaceus. Holotype (O-F21376). A. Mature ascomata. B. Detail of cortex. C. Ascoma in section. D. Peridial structure. E. Peridial structure between warts. F. Section of peridium showing warts (orange). G, H. Ascospores. Scale bars: A–C = 10 mm; D = 10 µm; E–H = 20 µm.
Fig. 8.

Type locality of Elaphomyces roseoviolaceus. Norway, Akershus, Frogn, Knardal Nature reserve, 3 Sep. 2013.
Fig. 9.

Elaphomyces granulatus and E. muricatus type specimens. A, B. Elaphomyces granulatus, epitype (GB-0147062). C, D. Elaphomyces muricatus, epitype (GB-0147063). E. Elaphomyces muricatus, holotype (UPS, F-127359) from Elias Fries’ herbarium. F. Type locality for epitype of Elaphomyces muricatus at Femsjö, Sweden, 15 Mar. 2014. Scale bars: 10 mm.
Synonyms: Lycoperdon cervinum L., Species plantarum 2: 1183. 1753.
Elaphomyces leucocarpus Vittad., Monographia Tuberacearum (Milano): 72. 1831.
Lectotype for Elaphomyces granulatus Fr. (designated here): Mougeot JB, Nestler CG, Schimper WP (1812). Stirpes cryptogamae vogeso-rhenanae; quas in Rheni superioris inferiorisque, nec non Vogesorum praefecturis, collegerunt J.B. Mougeot et C. Nestler. Fasc 3, No. 282, third basidioma from left (UPS F-708238). MBT389802.
Epitype (designated here): Sweden, Småland, Femsjö, mixed coniferous forest, 16 Mar. 2014 Lello (dog) & A. Molia, in herb. GB, GB-0147063*, barcode sequence KR029767, MBT389803.
Remarks: When Fries (1829) described Elaphomyces granulatus he referred to Lycoperdon cervinum of Linnaeus (1753) and cited several early illustrations, among them Lobelius (1581: 276; Tubera cervina), Bauhin (1651: 835; cervi boletus), Micheli (1729: tab. 99, fig. 4; Lycoperdastrum tuberosum), Nees von Esenbeck (1817: fig. 147; Tuber cervinum). There is no original type material available of E. granulatus and none of the illustrations referred to by Fries are accurate enough to enable us to clearly distinguish E. granulatus from related species. Fries also cited number 282 from the series of exsiccate published by Mougeot & Nestler (Mougeot & Nestler 1812; as Scleroderma cervinum). This exsiccate was distributed to a number of museums and other institutions throughout Europe, among them the herbaria in Lund, Stockholm, and Uppsala. The exsiccate copy in Uppsala contains a mixture of E. granulatus and E. hassiacus and the one in Lund consists of E. asperulus or a closely related species. The specimen of interest from the exsiccate copy in Stockholm could not be located. According to the nomenclatural code all cited specimens are syntypes. In this case it means that all exsiccate copies are syntypes and that a lectotype has to be chosen among them. Here we select one basidioma from the exsiccate copy in the Uppsala herbarium (UPS) that corresponds to the current interpretation of E. granulatus (Fig. 4). However, with the introduction here of the new species E. citrinopapillatus, which in our analyses is recovered as a sister taxon to E. granulatus, a unanimous morphological identification of old, single basidiomata cannot be guaranteed. Thus, the selected lectotype needs to be supplemented by an epitype. The epitype is newly collected in Femsjö, Fries’ mycological hunting-ground in southern Sweden (Fig. 9A–B).
Elaphomyces pusillus A. Molia & Sivertsen, sp. nov. Figs 5, 6. MycoBank MB833577.
Etymology: pusillus (small), referring to the size of the ascomata.
Typus: Norway, Nord-Trøndelag, Verdal, Inndalen, Elgstien at river Inna (WGS84: N63.70009 E11.89353), alt. 180 m, mesic, mossy Picea abies forest with some deciduous trees, with ferns and Vaccinium myrtillus in the field layer, 8 Sep. 2014, Lello (dog) & A. Molia, holotype in herb. O, O-F22174*, barcode sequence GenBank KR029761, isotype GB.
Ascomata globose to subglobose, 0.4–1.5(–1.8) cm, bright yellow (young) to dull brown (old; thus recalling E. asperulus). Peridial surface with angular and in relation to ascoma size rather big papillae, cream white to pale yellow. Odour weak, reminding of silver polish and reminiscent of the smell of E. asperulus and E. granulatus. Ascomata are often incrusted in a thick hyphal mat of cream-yellow to yellow hyphae with interwoven roots and debris. Peridium in section 0.7–1.3 mm thick, pink to slightly violaceous, darkening towards the gleba, homogeneous, composed of elongated, pseudoparenchymatic, thick-walled cells (1.5–2 µm in diam.), with red-brown pigments. Old and presumably overwintered specimens have a blueish halo close to the cortex. Outer peridial layer (cortex) with prominent, cubic to prismatic or conical, often cog-wheellike papillae, yellowish in section with a cellular structure, with cells of various shapes. Gleba initially greyish black, cottony, with age violaceous to brownish black, pulverulent. Pink tramal plates extend from the peridium into the gleba in young ascomata. Asci 8-spored. Ascospores globose, (20–)23–30(–33) µm (av. 26 µm, n = 100), when mature dark brown with an ornamentation of coalescing spines, forming a cracking pattern similar to that of E. asperulus. Spines 0.5–1(–1.5) µm high. Young spores are lighter brown and appear more regularly verrucose.
Ecology and distribution: Found with Picea abies and possibly Betula sp. on calcareous soil. The ascomata, which sometimes occur gregariously, develop in the upper soil, in a rather thick, cream-white hyphal mat. It is a rarely recorded species found on a few occasions in the boreal vegetation zone of Norway and Sweden.
Additional materials examined: Norway, Nordland, Rana, Jordbru, in Betula forest, 23 Sep. 1979, S. Sivertsen & B.K.P. Sveum, TRH 1273*; Nord-Trøndelag, Verdal, Inndalen, Elgstien at river Inna, in mesic, mossy Picea abies forest with some deciduous trees, ferns and Vaccinium myrtillus, 8 Sep. 2014, Lello (dog) & A. Molia, O-F22175*; Buskerud, Flesberg, Lyngdal, SE of Molia, in semi-rich Picea abies forest, 15 Aug. 2015, A. Molia, O-F22494, O-F22495. Sweden, Medelpad, Tuna, Runsvik, in rich Picea abies forest, 9 Sep. 2014, A. Molia, O-F22496; ibid., A. Molia, GB-0179907*.
Remarks: Elaphomyces pusillus is in many ways similar to E. asperulus, from which it differs in having extremely small-sized ascomata imbedded in a thick hyphal mat, and smaller ascospores. Elaphomyces pusillus has been found growing gregariously with more than 10 ascomata on the same spot. It was first collected in Rana (Nordland) in northern Norway by Sigmund Sivertsen and Bodil K. Sveum in 1979. The ascomata were on that occasion found loose on the ground, having been dug up by animals (likely by reindeer, Rangifer tarandus). Later records were exclusively made by trained dogs. Our molecular data confirm it as a distinct species, related to E. asperulus and E. roseoviolaceus, described below.
Elaphomyces roseoviolaceus A. Molia & E. Larss., sp. nov. Figs 7–8. MycoBank MB833578.
Etymology: from Latin roseus (pink/rose) and violaceus (violet), referring to the colour of the peridium in section.
Typus: Norway, Akershus, Frogn, Knardal NR (WGS84: N59.73711 E10.72019), in rich mixed forest with Pinus, Picea, Tilia, Corylus; under Pinus sylvestris, 3 Sep. 2013, Louise, Lello (dogs), K. Killingmo & A. Molia, holotype in herb. O, O-F21376*, barcode sequence KR029752.
Ascomata globose to subglobose, 1.3–2.5 cm (av. 1.6 cm, n = 23), pale sand brown to greyish brown, darker in old, overwintered specimens, with small flat and angular papillae. Ascomata are normally encrusted with soil particles and covered with a cream to dull yellow hyphal mat. Odour indistinct in the field, later (when enclosed in box) like freshly ground pepper. Peridium in section 1.4–2.1 mm thick, dark pink to bluish violaceous, most colourful towards the gleba; in periphery cream yellow. Peridium homogeneous in section with thick-walled (up to 2 µm), curved to elongated, sometimes rounded or angular cells, 8–40 µm. The cells contain pink to wine red pigments, particularly towards the cell wall. The cortex is 250–350 µm thick, with conical papillae unevenly distributed over the ascomatal surface. The papillae are yellow in section. Between the papillae there are pale yellow to colourless cells of two types: curved elongated thin-walled cells and thick-walled, irregularly arranged cells. The cortex is easily separated from underlying part of the peridium. Gleba black to violaceous when mature. Asci not seen. Ascospores (26–)27–33(–34) µm (av. 29.5 µm, n = 80), chestnut brown with darker ornamentation. Young spores have verrucose ornamentation whereas older spores have short (< 1 µm), coalescing spines, clustering in a patchy pattern.
Ecology and distribution: Occurs gregariously in the upper soil layer in calcareous, herb-rich woodlands with Picea abies and Pinus sylvestris. Hitherto known from four localities in southern Norway.
Additional materials examined: Norway, Telemark, Bamble, Langøya, in rich mixed coniferous forest, under Picea abies, 17 Oct. 2013, Lello (dog), T. Læssøe & A. Molia, O-F21324, O-F21429*; ibid., NE of Gjømle, rich mixed forest, under Picea abies, Lello (dog) & A. Molia, O-F22514; ibid., Røsskleiva, in rich mixed forest, under Picea abies, 20 May 2017, Lello (dog), I. Rokseth & A. Molia, O-F24144.
Remarks: Elaphomyces roseoviolaceus differs from E. asperulus by the more colourful, dark pink to violaceous peridium (in section), smaller ascomata and smaller spores. In the field, a thick cream or dull yellow mycelial mat is seen in the soil around the ascomata. In old and overwintered specimens, the peridium turns bluish-violaceous and the spores are slightly larger and have a more prominent ornamentation.
Elaphomyces muricatus Fr., Systema mycologicum (Lundiae) 3: 59. 1829. Fig. 9C–E.
Lectotype (designated here): Willdenow, C.L. (1787), Florae Berolinensis Prodromus, plate VII, fig. 19. MBT389804.
Epitype (designated here): Sweden, Småland, Femsjö, SW of Femsjö church (WGS84: N56.89087 E13.33069), in mixed woodland with Fagus sylvatica, Betula pendula and Pinus sylvestris, quite open area on a ridge, 15 Mar. 2014, Lello (dog) & A. Molia, in herb. GB, GB-0147062*, barcode sequence GenBank KR029730. MBT389807.
Remarks: In the Friesian herbarium at UPS there is a specimen (F-127359) that was collected at Femsjö (Sweden) and labelled Elaphomyces muricatus in Fries’ handwriting. The fruiting bodies show acute, pyramidal warts on the cortex surface and the peridium is clearly marbled in cross-section. However, only the peridium remains and no spores can be traced. Since the specimen is not dated it cannot be established if this material was available to Fries when describing the species and is thus not original material. In the protologue Fries refers to one illustration (in C.L. Willdenow Florae Berolinensis Prodromus, 1787) and this has to be selected as a lectotype. This type is here supplied with an epitype, based on newly collected and sequenced material from Femsjö.
Elaphomyces striatosporus Kers, Botaniska Notiser 133(2): 149. 1980. Fig 10.
Fig. 10.

Elaphomyces striatosporus Kers. A, C. Epitype (O-F238861). B, D. A rich collection with overwintered ascocarps. (O-F21368). E, F. Holotype (O-F72594). Scale bars: A, B, D, F = 10 mm; C = 20 µm.
Synonym: Elaphomyces papillatus var. striatosporus (Kers) A. Paz & Lavoise, Persoonia 38: 216. 2017.
Holotype: Norway, Oslo, ved Gausta, under Corylus, 22 Sep. 1952, F-E Eckblad, O-F72594.
Epitype (designated here): Norway, Oslo, Bygdøy, S. of Rodeløkken Café, under large Tilia sp. in a grazed field on calcareous soil, 21 Sep. 2012, Kokkos, Lello, Viktoria (dogs), K. Killingmo, M. Mowinkel-Amundsen & A. Molia, in herb. O, O-F245330*, barcode sequence GenBank KR029748. MBT389811.
Additional materials examined: Norway, Møre og Romsdal, Nesset, Eikesdal, under Rangåfjellet, in deciduous forest with Corylus avellana on semi-rich soil, 17 Sep. 2011, Kokkos, Viktoria (dogs), M. Jeppson, K. Killingmo & A. Molia, O-F21185*; Nord-Trøndelag, Inderøy, Råvika, in deciduous forest under Corylus avellana on rich soil, 21 Oct. 2011, Lello (dog) & A. Molia, O-F21184*; Oslo, Bygdøy, Klausåsen, S of Rodeløkken Café, in mixed deciduous forest under Corylus on calcareous ground, 21 Sep. 2012, Kokkos (dog), K. Killingmo & A. Molia, O-F245333*; Oslo, Hovedøya, deciduous forest with e.g. Corylus, Tilia, Quercus, Fraxinus, Betula on calcareous ground, 23 Sep. 2012, Kokkos, Lello (dogs), K. Killingmo & A. Molia, O-F245337*; Østfold, Jeløy, Albyskogen, in mixed forest with Corylus, Quercus, Populus, Betula, and Picea, 17 Mar. 2014, Lello (dog), A. Fæste & A. Molia, O-F21368.
Remark: We made several attempts to generate an ITS sequence from the holotype but failed. For this reason, we consider it desirable to select an epitype using recently collected and sequenced material from the vicinity of the type locality.
DISCUSSION
The phylogeny and taxonomy of Elaphomyces in Europe was recently treated in detail by Paz et al. (2017). Twenty-six species were identified and an intrageneric division was established, supported by analyses of ITS and LSU sequence data. Four major clades were identified and classified as sections, viz. sect. Elaphomyces, Ascoscleroderma, Ceratogaster and Malacodermei. Representatives of the sections Elaphomyces, Ceratogaster and Malacodermei are currently known to occur in North Europe but only section Elaphomyces is treated in this paper. Paz et al. (2017) divided section Elaphomyces in three subsections: Elaphomyces, Muricati and Papillati. The species recovered in our analyses (Fig. 1) are briefly discussed below.
Subsection Elaphomyces
The main morphological character of subsection Elaphomyces is the presence of a homogenous (not marbled) peridium in section. Three European species belong to this subsection according to Paz et al. (2017): E. asperulus, E. granulatus and E. hassiacus. The former two species are well known and abundant in North Europe, whereas the latter one has no current records.
During our fieldwork we noticed the presence of morphologically deviating collections that could not be referred to the above species, but which clearly belonged in this subsection. These observations were later supported by molecular data (Fig. 1). The three new species, E. citrinopapillatus, E. pusillus and E. roseoviolaceus form ectomycorrhiza with Picea abies and are characteristic elements in hemiboreal-boreal coniferous woodlands on more or less calcareous soil. A sister species of E. pusillus was also recovered in the phylogenetic analyses (Fig. 1). It has an almost identical morphology and similar habitat preferences and is here treated as E. cf. pusillus, since we need additional material for a formal description. The new species are currently known from Norway, and E. pusillus in addition from Sweden.
Subsection Muricati
Subsection Muricati includes species with a peridium that is marbled when observed in section. In the phylogeny (Fig. 1) sequence data of the epitype of E. muricatus, designated above, as well as the epitypes of E. muricatus var. variegatus and E. muricatus var. reticulatus are marked in bold-face type. Although there are only a few base pair differences in ITS sequences compared with the epitype of E. muricatus, the two varieties can be distinguished by morphology (Paz et al. 2017). A sister taxon, here provisionally treated as E. cf. muricatus (Fig. 1), was recovered in the phylogenetic analyses. It is represented by three molecularly identical collections from Norway. In morphology it is close to E. muricatus var. muricatus but more specimens must be studied before it can be formally described. Paz et al. (2017) described Elaphomyces violaceoniger, a species characterized by a dark purplish peridium, and reported a single record from Norway. We have not been able to confirm this record and currently we do not accept E. violaceoniger as a member of the Nordic funga. Elaphomyces quercicola, formerly E. muricatus f. quercicola, was elevated to species rank by Paz et al. (2017); so far, no records from North Europe have been confirmed.
Another species in the muricatus-group is E. barrioi, described by Paz et al. (2017) with a holotype from Spain. It is likely to be confused with E. muricatus but careful observation of peridial features should make it possible to separate the two (see identification key below). It forms ectomycorrhiza with deciduous trees and has several records from southern Europe and is also present in southern Norway (Fig. 1). A sister taxon, collected in Norway and clearly separated by molecular data, is provisionally treated as E. cf. barrioi. Since only one small collection is known a formal description will have to await additional gatherings.
Elaphomyces decipiens also belongs to subsection Muricati. The species was neotypified with Spanish material and sequence data of the neotype generated (Paz et al. 2017). In our analyses, collections from Norway, and Sweden determined as E. decipiens based on morphology, form a sister clade to South European specimens (Fig. 1). The clades differ in ITS sequence data by four substitutions and three insertion/deletion events and should likely be regarded as two separate species and are in need of further attention (cf. Jeppson & Molia 2015).
Subsection Papillati
Subsection Papillati includes species with striate spores. Paz et al. (2017) divided Elaphomyces papillatus into three varieties, var. papillatus, var. striatosporus and var. sulphureopallidus. Of these only var. striatosporus occurs in northern Europe. The ITS sequence of this variety differs from var. papillatus by 2–4 insertions. In morphology E. striatosporus is best distinguished from E. papillatus by a less warty peridial surface and a more pronounced striation of the spore wall. Var. striatosporus was originally described on species level based on material collected by Finn-Egil Eckblad from Oslo, Norway, 1952 (Kers 1980). As this name is in current use in Norway and Sweden, and as the fungus can be distinguished by both morphology and ITS sequence data, we currently prefer to treat it on species level, as a northern taxon, closely related to the South European E. papillatus. As we were not able to generate ITS sequence data from the holotype material of E. striatosporus (O-F72594; Fig. 10E–F), the species is in this paper epitypified with newly collected material from Oslo.
Kers (1980) classified E. striatosporus in subgenus Malacoderma. The molecular analyses (Paz et al. 2017, this study) confirm its position among the "brown species" in section Elaphomyces and its peridium is in fact brown, particularly apparent in older stages.
In this paper we show that nine species belonging to Elaphomyces section Elaphomyces are present in northern Europe. This is more than a doubling of the number keyed out by Eckblad, Lange & Kers in the flora Nordic Macromycetes (Hansen & Knudsen 2000). In addition, our phylogenetic analyses of DNA sequences revealed the presence of genetically divergent specimens that could represent another four taxa (Fig 1.) It is obvious that much fieldwork remains in order to give a realistic picture of the diversity and the true distributions of Elaphomyces species in northern Europe. This is especially true for Denmark, where almost no collecting taking the advantage of trained dogs has taken place so far. Although Elaphomyces specimens often are found somewhat deeper in the ground than other hypogeous fungi, trained dogs are very efficient in locating the ascomata. For the prospective truffle-hunter Elaphomyces has the advantage over many other truffles in being present and detectable all year around.
Elaphomyces spp. are important contributors to forest ecosystem functioning, forming ectomycorrhiza with forest trees (Bird & McCleneghan 2005). Three species from subsection Elaphomyces (viz. E. asperulus, E. granulatus and E. muricatus) have abundant records from North European hemiboreal and boreal coniferous forests according to our studies and to herbarium data and records in national databases. However, species identifications of older records are ambiguous due to the presence of previously unknown and morphologically similar species. The herein proposed new species occur mainly in old-growth herb-rich Picea forests on calcareous ground and may turn out to be useful indicators for this type of habitat that is under a continuous threat by modern forestry.
Key to North European species of Elaphomyces - all sections
1. With black, brownish black to blueish black or greyish black cortex, or at least black peridial spines ............................................... 2
1. With brown or brownish cortex, never with black peridial spines .................................................................................................... 11
2. Cortex warty, or partly warty, with acute or blunt warts ..................................................................................................................... 3
2. Cortex smooth or nearly smooth ......................................................................................................................................................... 7
3. Ascospores with striate-ridged ornamentation ................................................................................................................................... 4
3. Ascospores non-striate ........................................................................................................................................................................ 5
4. Ascomata 10–35 mm; peridial surface minutely and densely warty, very dark brown to black; ascospores 16–22 µm, ornamentation with sharp ridges ................................................................................................................... E. virgatosporus
4. Ascomata usually < 10 mm, when young often wrapped in a thick white to cream-coloured mycelial felt; peridium scurfy to finely papillate, in young ascomata blackish blue, with age bluish grey to chestnut brown; ascospores 11–18 µm, ornamentation with blunt ridges ................................................................................ E. striatosporus
5. Peridium with blackish spines penetrating the brown peridial surface ............................................................................. E. aculeatus
5. Peridium different ................................................................................................................................................................................ 6
6. Ascospores with coalescent ornamentation of low and dense warts ................................................................................... E. moretti
6. Ascospores with small spines/warts, forming a dotted pattern on the surface .................................................................... E. leveillei
7. Ascospores >25 µm; ascomata 20–50 mm, with patches of a blue green mycelial felt on the peridium; peridial surface black, almost smooth .................................................................................................................. E. maculatus
7. Ascospores < 25 µm ............................................................................................................................................................................. 8
8. Ascomata greyish; mature gleba whitish-pinkish to dirty grey; ascospores 32–36 µm, pale-coloured ............................... E. septatus
8. Ascomata blackish, ascospores darker, gleba different ....................................................................................................................... 9
9. Ascomata with yellow papillae, breaking through the blackish peridium .............................................................. E. citrinopapillatus
9. Ascomata smooth .............................................................................................................................................................................. 10
10. Ascomata small, normally between 5–15 mm, globose to ± pyriform, some with prominent depressions or ± flattened; ascomata gregarious........................................................................................................................................... E. anthracinus
10. Ascomata normally > 20 mm; spores often angular; ascomata solitary ................................................. E. anthracinus f. talosporus
11. Inner peridium marbled in section .................................................................................................................................................. 12
11. Inner peridium uniform in section ................................................................................................................................................... 14
12. Peridial surface with blunt and flat warts, pale brown to dirty brown; a thick dull yellow hyphal mat covers the ascomata; ascospores 19–28 µm with a dense cover of curved, rod-like spines ................................................................ E. cf. decipiens
12. Peridial surface with ± acute warts .................................................................................................................................................. 13
13. Peridial surface with prominent, acute warts, 1(–2) per mm; ascospores 18–21 µm, with thin, curved and rod-like spines ................................................................................................................................................................................. E. muricatus
13. Peridial surface with small, truncate warts, 2(–3) per mm; ascospores 19–24 µm, with thick, rod-like spines with confluent apices, forming irregular meshes ............................................................................... E. barrioi
14. Cortex surface blackish brown with yellow warts; ascomata 6–20 mm; ascopores 23–28 µm, with long, easily separable spines (similar ornamentation to other species in the granulatus-group) ....................... E. citrinopapillatus
14. Cortex surface brownish .................................................................................................................................................................. 15
15. Ascomata small, normally 4–15(–18) mm, often encrusted by a thick hyphal mat of cream-coloured to clearly yellow hyphae with interwoven roots and debris; ascospores 23–30 µm.................................................................. E. pusillus
15. Ascomata larger, normally > 20 mm in diam. .................................................................................................................................. 16
16. Peridium with pink to violaceous tints; ascomata 13–25 mm; peridial surface light brown with small, flat papillae; peridium strongly coloured, dark pink to violaceous in section; ascospores 26–31 µm with low, spiny verrucae, with age coalescing and forming small isolated groups (cheetah pattern), ascomata gregarious ......................................................................................................................................................................... E. roseoviolaceus
16. Peridium dull pink, cream coloured, rarely bluish; ascomata normally larger ................................................................................ 17
17. Cortex ochre yellow-brown; peridium whitish to yellowish white in section; ascomata 15–40 mm; ascospores 20–29 µm, with an ornamentation of isolated spines ....................................................................... E. granulatus
17. Cortex dull brow; peridium with a pinkish tint in section, sometimes with a blue halo (in old/overwintered specimens); ascomata normally 30–40 mm; ascospores 25–38 µm with low, dense verrucae, with age forming a patchy pattern with groups of coalescing spines divided by narrow paths (giraffe-pattern) ......................................................... E. asperulus
ACKNOWLEDGEMENTS
Curators of the herbaria C/CP, O, S, TRH, UPS are gratefully acknowledged for arranging loans. P-A Moreau, A. Paz and C. Lavoise are thanked for valuable discussions, collections, and for input to the investigation of the Mougeot & Nestler exsiccate. We are especially grateful for the help from Svengunnar Ryman to interpret the nomenclatural code and for comments and discussions related to the typification of Friesian material. Financial support was received from The Norwegian Biodiversity Information Centre (ArtsDatabanken; ref. nr 67–10), The Norwegian Barcode of Life Network (NorBOL), The Swedish Taxonomy Initiative, (ArtDatabanken), and Carl Stenholm’s foundation.
Conflict of interest: The authors declare that no known conflicts of interests exists.
REFERENCES
- Bauhin J. (1651). Historia plantarum universalis, nova, et absolutissima, cum consensus et dissensucirca eas. Vol. 3 Yverdon, France. [Google Scholar]
- Bird C, McCleneghan C. (2005). Morphological and functional diversity of ectomycorrhizal fungi on Roan Mountain (NC/TC). Southeastern Naturalist 4: 121–132. [Google Scholar]
- Buyck B, Hosaka K, Masi S, et al. (2016). Molecular analyses of first collections of Elaphomyces Nees (Elaphomycetaceae, Eurotiales, Ascomycota) from Africa and Madagascar indicate that the current concept of Elaphomyces is polyphyletic. Cryptogamie Mycologie 37: 3–14. [Google Scholar]
- Castellano MA, Trappe JM, Vernes K. (2011). Australian species of Elaphomyces (Elaphomycetaceae, Eurotiales, Ascomycota). Australian Journal of Systematic Botany 24: 32–57. [Google Scholar]
- Castellano MA, Beaver RE, Trappe JM. (2012a). Sequestrate fungi of New Zealand: Elaphomyces (Ascomycota, Eurotiales, Elaphomycetaceae). New Zealand Journal of Botany 50: 423–433. [Google Scholar]
- Castellano MA, Dentinger BTM, Séné O, et al. (2016). New species of Elaphomyces (Elaphomycetaceae, Eurotiales, Ascomycota) from tropical rainforests of Cameroon and Guyana. IMA Fungus 7: 59–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano MA, Guerrero GC, Jiménez JG, et al. (2012b). Elaphomyces appalachiensis and E. verruculosus sp. nov. (Ascomycota, Eurotiales, Elaphomycetaceae) from eastern North America. Revista Mexicana de Micología 35: 17–22. [Google Scholar]
- Castellano MA, Henkel TW, Miller SL, et al. (2012c). New Elaphomyces species (Elaphomycetaceae, Eurotiales, Ascomycota) from Guyana. Mycologia 104: 1244–1249. [DOI] [PubMed] [Google Scholar]
- Castellano MA, Stephens RB. (2017). Elaphomyces species (Elaphomycetaceae, Eurotiales) from Bartlett Experimental Forest, New Hampshire, USA. IMA Fungus 8: 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, et al. (2012). Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana E. (1909). Ricerche intorno di alcune species del genere Elaphomyces Nees (E. variegatus – E. granulatus e affini). Memorie della Reale accademia delle scienze di Torino ser. II 59: 89–108, pl 1–2. [Google Scholar]
- Fries EM. (1829). Systema mycologicum III Gryphiswaldae, Germany. [Google Scholar]
- Gardes M, Bruns TD. (1993). ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizas and rusts. Molecular Ecology 2: 113–118. [DOI] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. (1995). Development of primer sets designated for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen L, Knudsen H. (2000). Nordic Macromycetes vol. 1 Ascomycetes. Nordsvamp, Denmark. [Google Scholar]
- Hopple JS Jr, Vilgalys R. (1999). Phylogenetic relationships in the mushroom genus Coprinus and dark-spored allies based on sequence data from the nuclear gene coding for the large ribosomal subunit RNA: divergent domains, outgroups, and monophyly. Molecular Phylogenetics and Evolution 13: 1–19. [DOI] [PubMed] [Google Scholar]
- Index Fungorum (2018). http://www.indexfungorum.org. [Google Scholar]
- Jeppson M, Molia A. (2015). Två hypogeiska svampar nya för landet – Elaphomyces decipiens och Hymenogaster bulliardii. Svensk Mykologisk Tidskrift 36: 36–41. [Google Scholar]
- Katoh K, Standley DM. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kers LE. (1980). A new species of Elaphomyces Nees ex Fr. subgen. Malacoderma Vitt. Botaniska Notiser 133: 149–153. [Google Scholar]
- Larsson E, Örstadius L. (2008). Fourteen coprophilous species of Psathyrella identified in the Nordic countries using morphology and nuclear rDNA sequence data. Mycological Research 112: 1165–1185. [DOI] [PubMed] [Google Scholar]
- Ławrynowicz M. (1988). Grzyby (Mycota). Workowce (Ascomycetes). Jeleniakowe (Elaphomycetales). Truflowe (Tuberales). Flora Polska 18. Polska Akademia Nauk, Poland. [Google Scholar]
- Ławrynowicz M. (1989). Chorology of the European hypogeous Ascomycetes. I. Elaphomycetales. Acta Mycologica 25: 3–41. [Google Scholar]
- Linnaeus C. (1753). Species plantarum. Tomus II. Holmiae, Sweden. [Google Scholar]
- Lobelius M. (1581). Plantarum seu stirpium icons. Antwerpiae, Netherlands. [Google Scholar]
- Micheli PA. (1729). Nova plantarum genera. Florentiae, Italy. [Google Scholar]
- Mougeot JB, Nestler CG, Schimper WP. (1812). Stirpes cryptogamae vogeso-rhenanae; quas in Rheni superioris inferiorisque, nec non Vogesorum praefecturis, collegerunt J.B. Mougeot et C. Nestler. Fasc 3. 201–300. Bruyerii Vogesorum (Vivot), France. [Google Scholar]
- Nees von Esenbeck CG. (1817). Das System der Pilze und Schwämme. Würzburg, Germany. [Google Scholar]
- Nees von Esenbeck CG, Nees von Esenbeck TFL. (1820). Jacob Boltons Geschichte der Merkwürdigsten Pilze IV. Synopsis generum plantarum mycetoidarum. Berlin, Germany. [Google Scholar]
- Nylander JAA. (2004). MrModeltest version 2. – Program distributed by the author. https://github.com/nylander/MrModeltest2 [Google Scholar]
- Paz A, Bellanger JM, Lavoise C, et al. (2017). The genus Elaphomyces (Ascomycota, Eurotiales): a ribosomal DNA-based phylogeny and revised systematics of European “deer truffles”. Persoonia 38: 197–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds HT. (2011). Systematics, Phylogeography and Ecology of Elaphomycetaceae. PhD dissertation. Biology, Duke University, USA. [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2012). MrBayes 3.2, efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. (2003). PAUP*. Phylogenetic Analysis Using Parsimony (* and other methods). Version 4. Sinauer, USA. [Google Scholar]
- Tulasne L-R, Tulasne C. (1841). Sur le genre Elaphomyces, et description de quelques espèces nouvelles. Annales des Sciences Naturelles; Botanique, sér. 2, 16: 5–27, pl 1–4. [Google Scholar]
- Tulasne L-R, Tulasne C. (1851). Fungi hypogaei. Histoire et monographie de champignons hypogés. F. Klinckseick, Paris. [Google Scholar]
- Vittadini C. (1831). Monographia tuberacearum. Typographia F. Rusconi, Milano. [Google Scholar]
- Vittadini C. (1842). Monographia Lycoperdineorum. Augustae Taurinorum ex officina Regia, Torino. [Google Scholar]
- White TJ, Bruns T, Lee L, et al. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols, A guide to Methods and Applications (Innis MA, Gelfand DH, Sininski JJ, et al., eds.). Academic Press, USA: 315–322. [Google Scholar]
- Zhang BC. (1991). Revision of Chinese species of Elaphomyces (Ascomycota, Elaphomycetaceae). Mycological Research 95: 973–985. [Google Scholar]
- Zhang BC, Minter D. (1989). Elaphomyces spinoreticulatus sp. nov. with notes on Canadian species of Elaphomyces. Canadian Journal of Botany 67: 909–914. [Google Scholar]


