Figure 7.
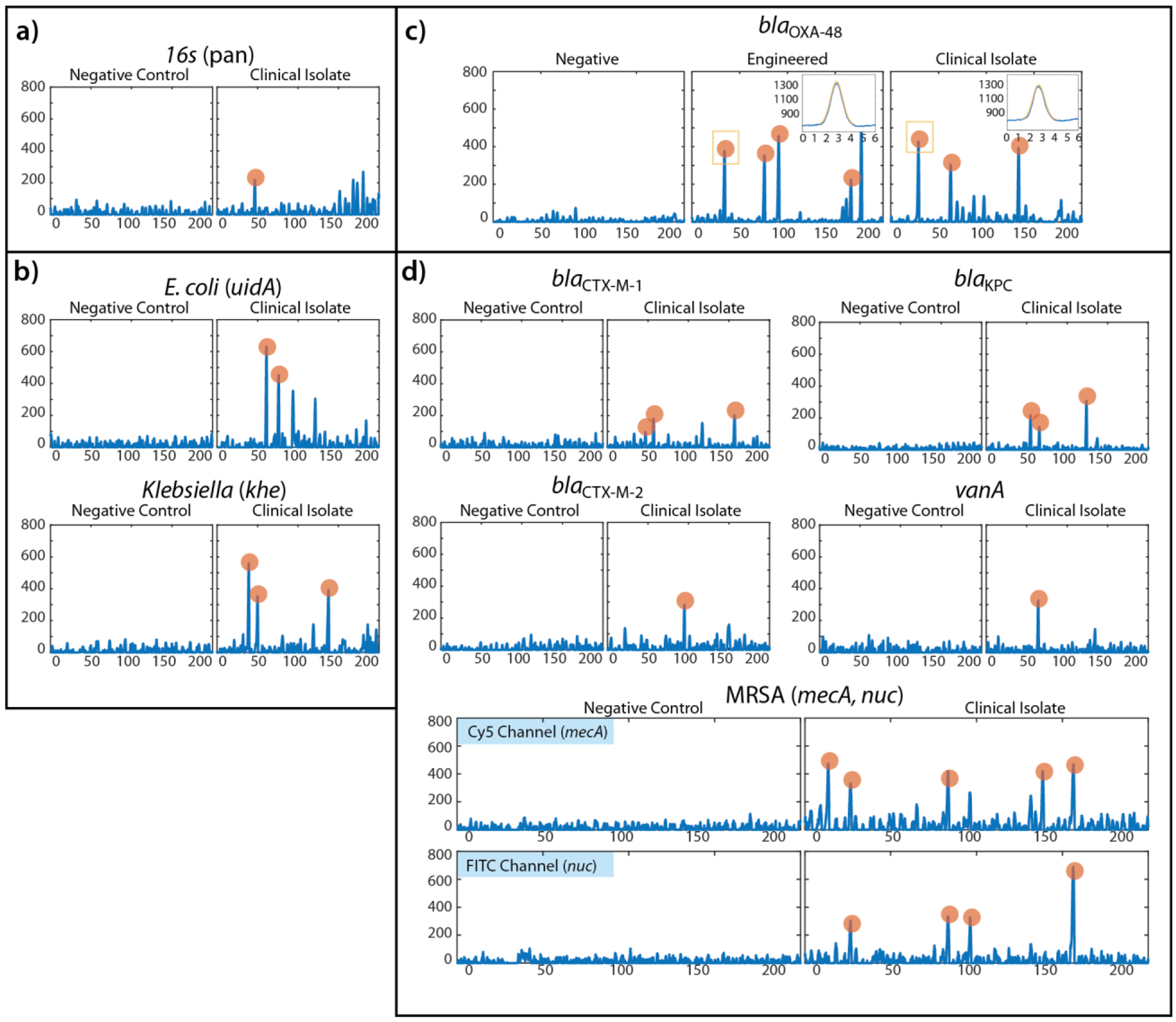
Demonstration of wide applicability of IC3D blood ddPCR technology for the rapid diagnosis and mangement of BSI and antibiotic resistance. a) Pan-bacteria detection with 16s gene panel. b) Species-identification panel including E. coli- (uidA) and Klebsiella- (khe) specific genetic markers. c) Representative example of blaOXA-48 as a model using internal negative control and positive control (engineered bacteria) along with clinical isolate. Inset panels for engineered control and clinical isolates demonstrate representative peaks identified with shape-fitting algorithm (X-axis = 6ms of data at 64kHz, Y-axis = raw signal intensity in mV). d) Antibiotic resistance genetic panel including blaCTX-M-1, blaCTX-M-2, vanA, and blaKPC, and MRSA (nuc, mecA). Each panel is a representative portion of the raw fluorescence signal recorded by the particle counter instrument. X-axis = 200ms of data at 64kHz, Y-axis = offset signal amplitude in mV). Orange highlighted peaks denote signal fluctuations that match the stringent shape-fitting criteria consistent with positive, fluorescent target-containing droplets.
