Fig. 2.
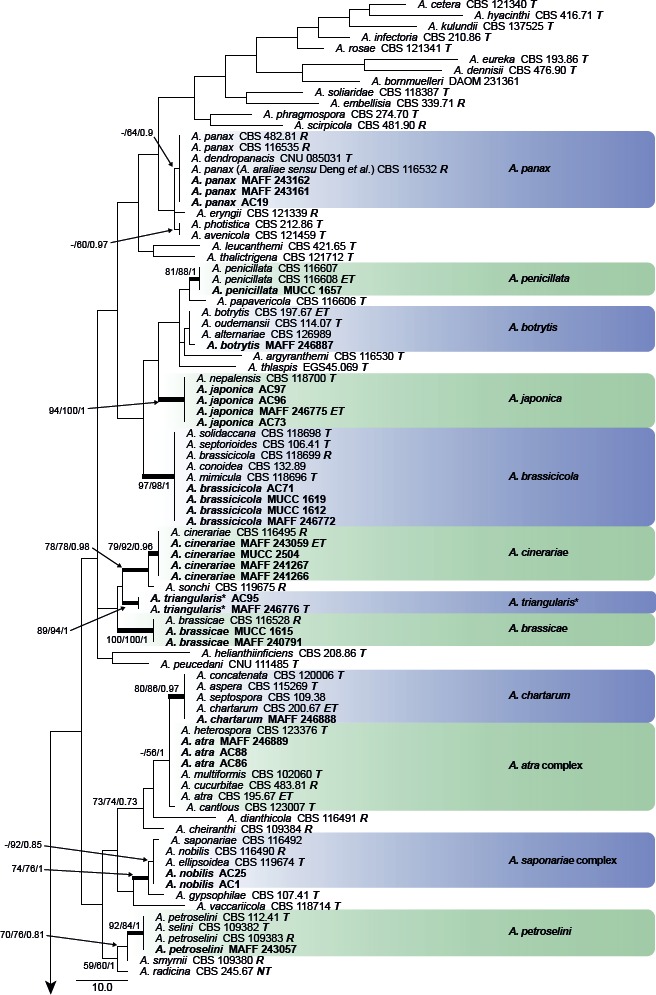
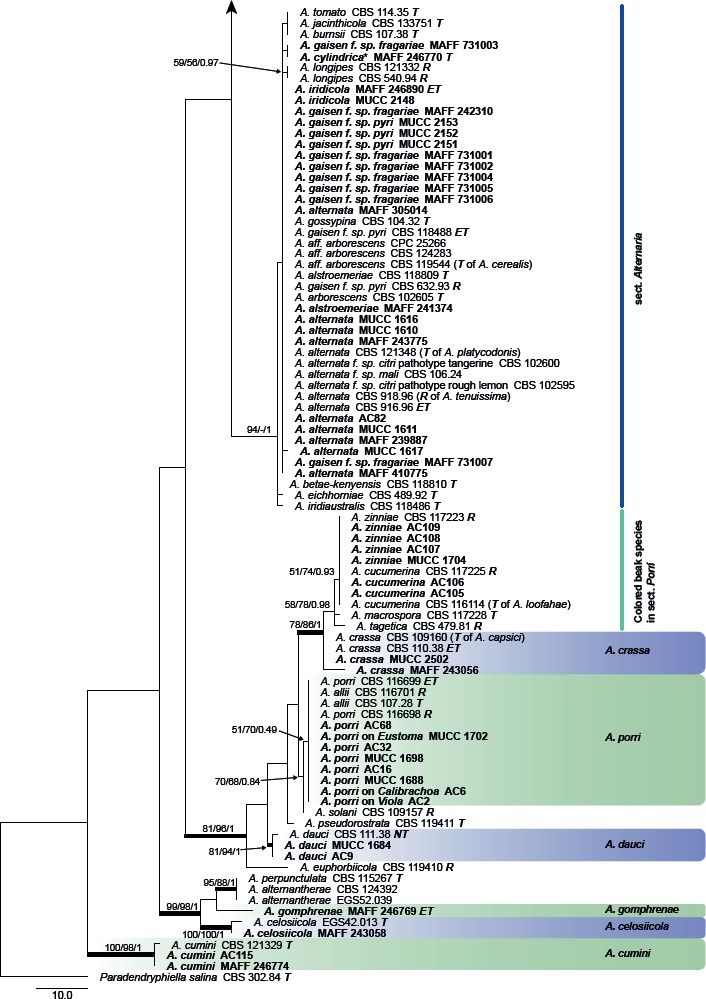
Phylogenetic tree generated from maximum parsimony (MP) analysis based on the ITS sequences from Japanese Alternaria isolates. The tree was rooted to Paradendryphiella salina (CBS 302.84). MP and RAxML maximum likelihood (ML) bootstrap values and Bayesian posterior probabilities (PP) are given near branches (MP/ML/PP). Thickened nodes indicate significant support by MP/ML/PP (> 60/60/0.96). Tree length = 600, consistency index = 0.433, homoplasy index = 0.567, retention index = 0.881, and rescaled consistency index = 0.382. The scale bar indicates the number of nucleotide substitutions. Japanese Alternaria isolates examined are indicated in bold, and the statuses of reference isolates are indicated in bold and italic. T: ex-type, NT: ex-neotype, ET: ex-epitype, R: representative strain assigned by Simmons (2007). Asterisks indicate novel taxa proposed in the taxonomy section.
