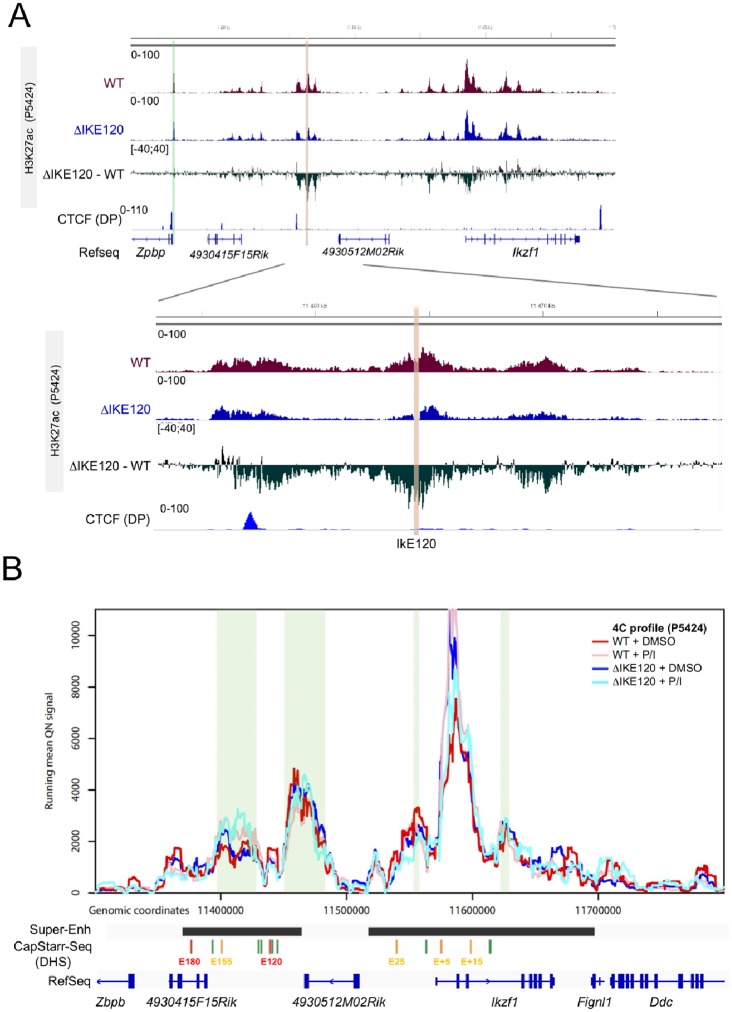
Fig 4. Epigenomic impact of IkE120 deletion.
(A) The H3K27ac ChIP-seq at the Ikzf1 locus (top) and around the IkE120 enhancer (bottom) in wt and ΔIkE120 P5424 cells are shown as individual tracks and as the differential signal between wt and ΔIkE120 cells. The genomic track of CTCF ChIP-seq in primary DP thymocytes is also shown. H3K27ac The IkE120 deleted region is highlighted in red and the promoter region of the neighbor Zpbp gene is highlighted in green. (B) Running mean (window of 21 fragments), quantile normalized 4C-seq profiles are shown from the Ikzf1 promoter bait for wt unstimulated (red), wt stimulated (pink), ΔIkE120 unstimulated (blue) and ΔIkE120 stimulated P5424 cells. Locations of genes and the six regions with enhancer activity in CapStarr-seq are shown below the plot. Conserved called interactions are highlighted in green.