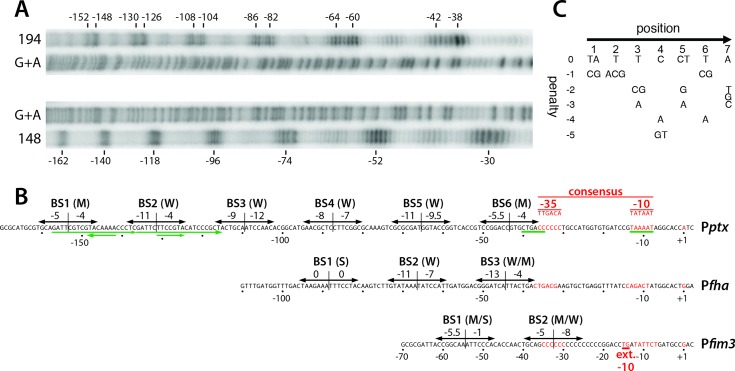
Fig 4. Details of BvgA-binding to the pertussis toxin promoter.
A. BvgAΔ127–129 derivatives in which both naturally occurring cysteine residues had been replaced by alanine and in which either the valine at position 148 or the threonine at position 194 had been replaced with cysteine, were subjected to derivatization with FeBABE and used to reveal the locations of BvgA-binding to the ptx promoter using methods previously described for a similar analysis of the fha promoter [12]. The protein modified at the 148 position produces cleavages at the outer boundaries of a bound dimer of BvgA~P, while the 194 derivative produces closely spaced cleavages corresponding to the location of the inter-monomer interface. Maxam and Gilbert A + G reactions of the same 32P end-labelled Pptx DNA fragment were run in parallel for orientation. B. DNA sequences of the ptx promoter showing the sites of binding of BvgA~P derived from the analysis in panel A. In addition, each heptameric half-site has been scored according to the algorithm presented in panel C, with the scores given above the arrows indicating the binding half-sites. Nucleotides in red indicate core promoter elements, with the consensus sequence shown above. In a similar fashion, Pfha and Pfim3 are shown for comparison. Green arrows below the Pptx sequence indicate two 21 bp imperfect direct repeats and two inverted heptameric imperfect repeats previously cited as potential BvgA binding sites. C. Algorithm for predicting binding strength of BvgA-binding half-sites. This algorithm was derived from a study examining the ability of systematically mutated derivatives of the Pfha primary binding site to bind BvgA~P and to activate transcription [14].