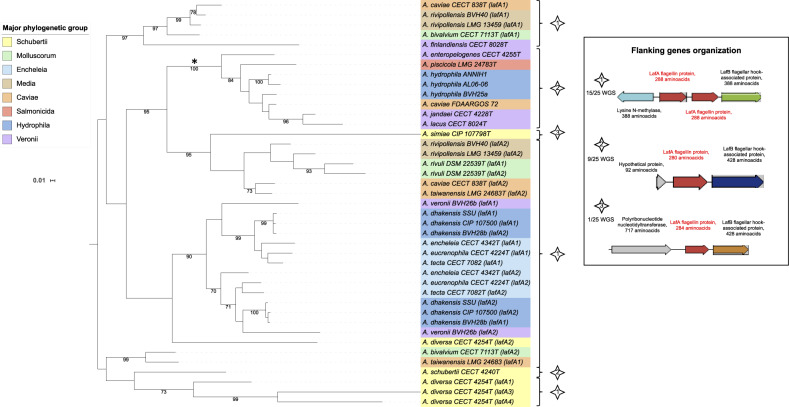
Fig. 4.
—ML tree based on lafA gene sequences (906 nt) reconstructed using the TIM model plus gamma distribution and invariant sites as a substitution model from 41 complete lafA nucleotide sequences. Each different copy found in a genome (or a strain) was arbitrarily numbered (lafAx). The same lafA numbering but in two different species corresponded to different lafA sequences. The clade gathering most of the genomes with a monocopy of lafA is indicated by an asterisk (*). The numbers at the nodes are support values estimated with 100 bootstrap replicates. Only bootstrap values ≥70 are indicated. The major phylogenetic group of each strain is indicated by colored ranges on strain labels. The type of genetic organization of the flanking genes shown in the inserted box is indicated for each strain with a numbered star. Abbreviation: WGS, whole-genome sequences.