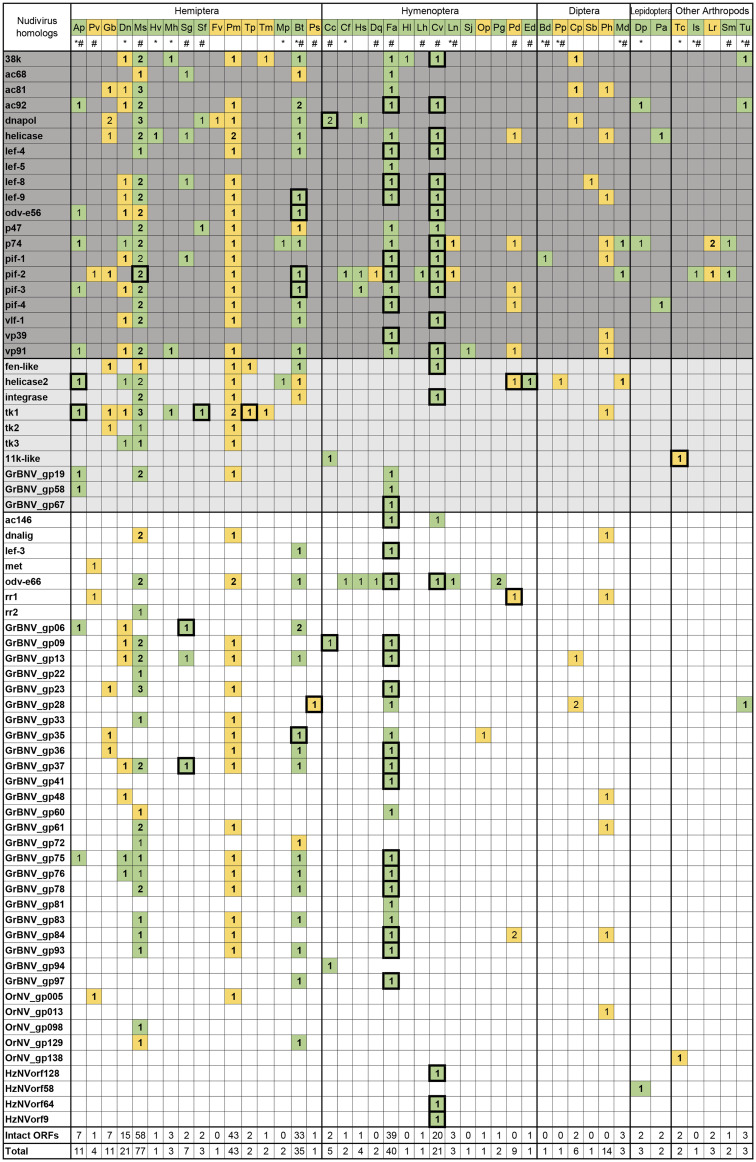
Fig. 1.
—Nudivirus-like genes identified in arthropod genomes. Copy numbers for each nudivirus homolog are indicated. Core genes conserved between baculoviruses and nudiviruses are shown in dark gray, and nudivirus-specific core genes are shown in light gray. Sequences with or without flanking host sequences were colored in green and yellow, respectively. Ap, Acyrthosiphon pisum; Pv, Pachypsylla venusta; Gb, Gerris buenoi; Dn, Diuraphis noxia; Ms, Melanaphis sacchari; Hv, Homalodisca vitripennis; Mh, Maconellicoccus hirsutus; Sg, Schizaphis graminum; Sf, Sipha flava; Fv, Ferrisia virgate; Pm, Paracoccus marginatus; Tp, Trionymus perrisii; Tm, Trabutina mannipara; Mp, Myzus persicae; Bt, Bemisia tabaci; Ps, Philaenus spumarius; Cc, Cephus cinctus; Cf, Camponotus floridanus; Hs, Harpegnathos saltator; Dq, Dinoponera quadriceps; Fa, Fopius arisanus; Hl, Habropoda laboriosa; Lh, Linepithema humile; Cv, Cotesia vestalis; Ln, Lasius niger; Sj, Synergus japonicas; Op, Ormyrus pomaceus; Pg, Pseudomyrmex gracilis; Pd, Polistes dominula; Ed, Euglossa dilemma; Bd, Bactrocera dorsalis; Pp, Phlebotomus papatasi; Cp, Condylostylus patibulatus; Sb, Sphyracephala brevicornis; Ph, Phortica variegate; Md, Mayetiola destructor; Dp, Danaus plexippus; Pa, Papilio glaucus; Tc, Triops cancriformis; Is, Ixodes scapularis; Lr, Loxosceles reclusa; Sm, Stegodyphus mimosarum; and Tu, Tetranychus urticae. *EST data available; #TSA data available. Boxes surrounding the numbers indicate that gene expression was detected. Sequences with intact ORFs were indicated by boldface.