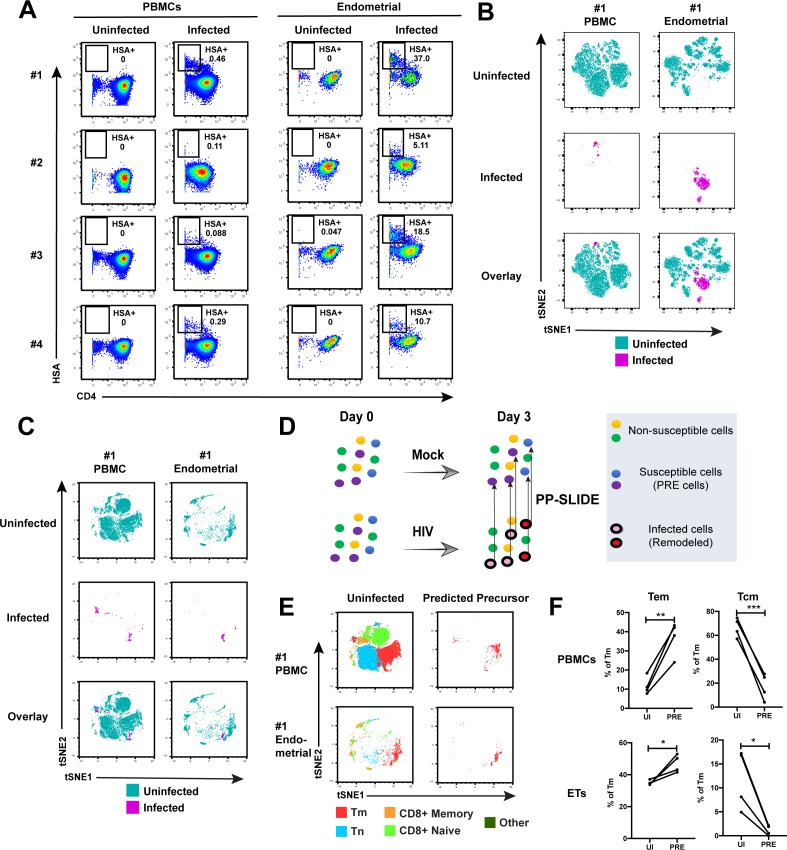
Figure 2. Comparison of HIV-susceptible cells in unstimulated PBMCs and endometrium.
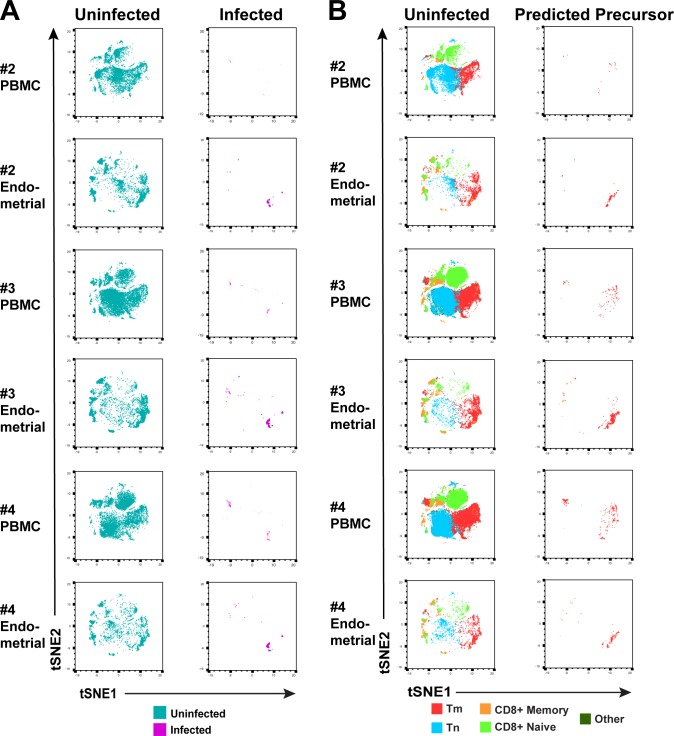
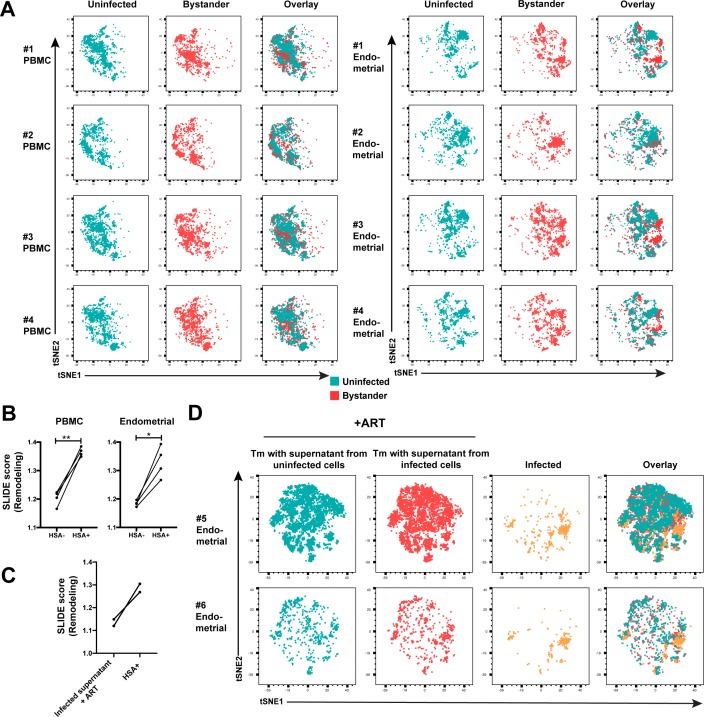
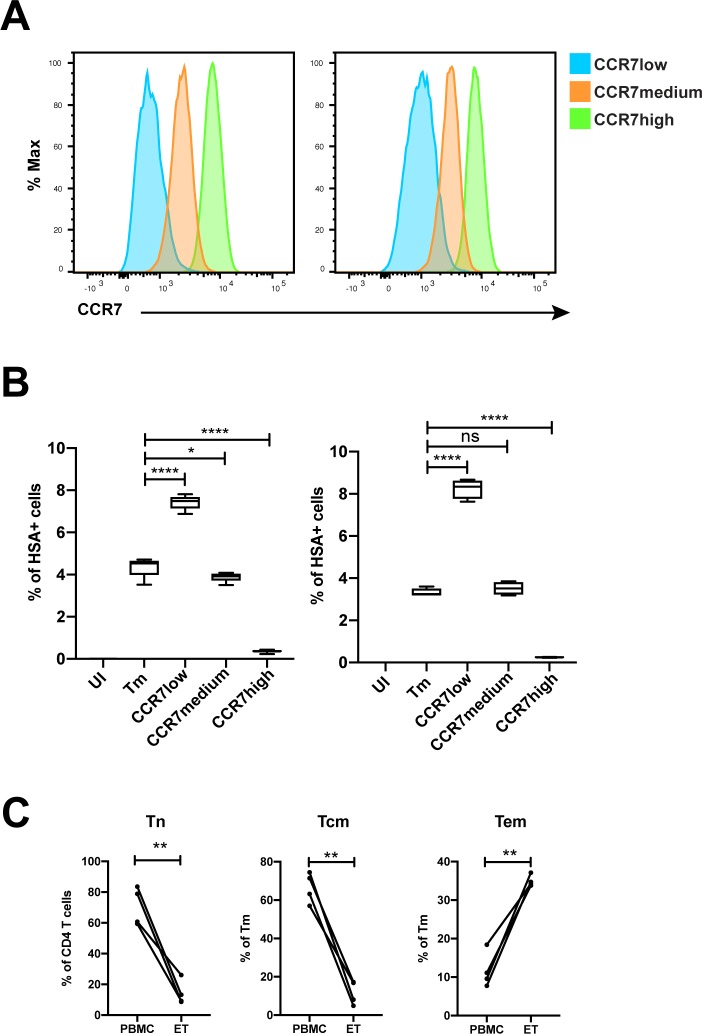
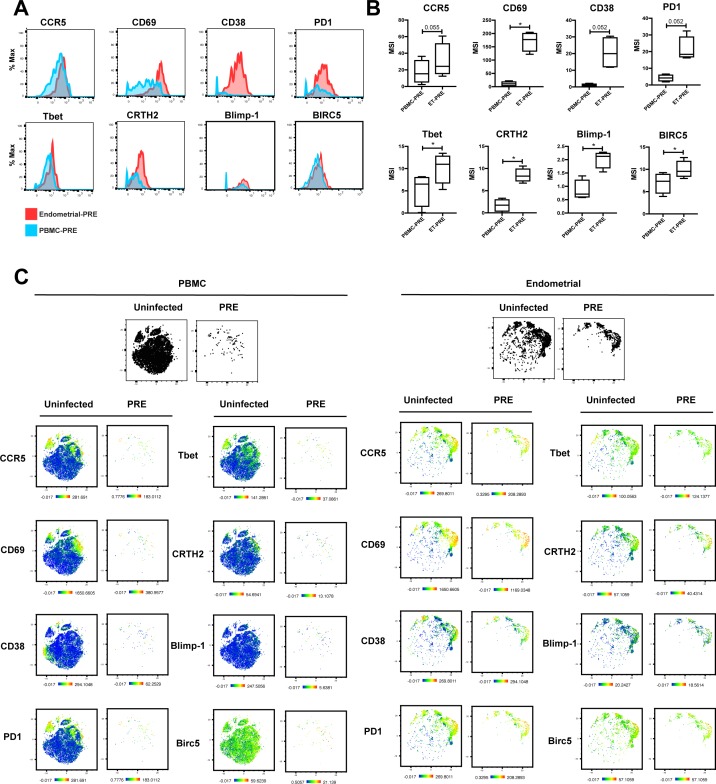
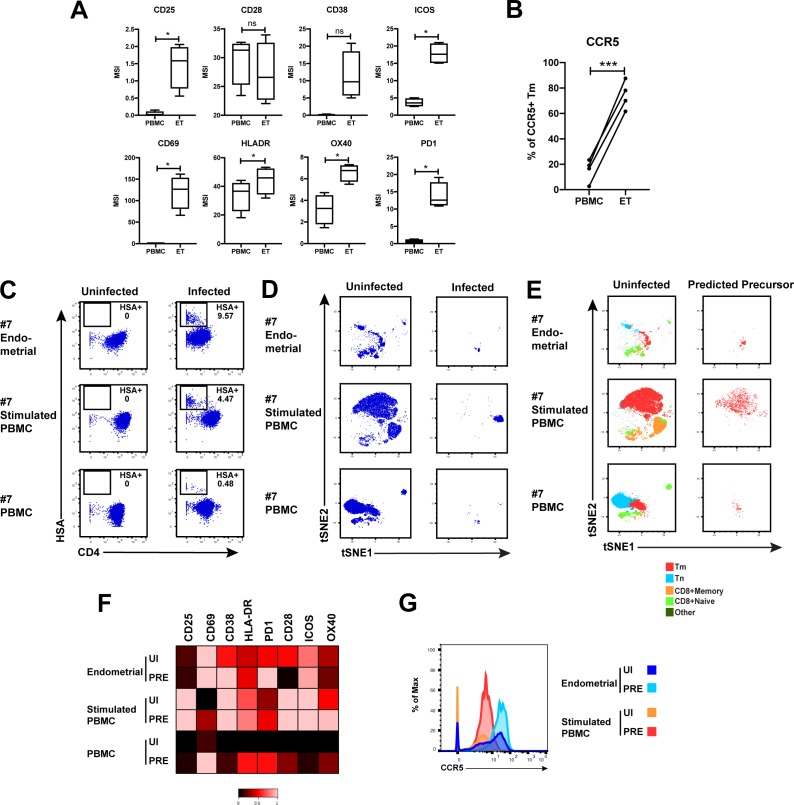
(A) Paired unstimulated PBMCs and endometrial cells from 4 donors were mock-treated or inoculated with F4.HSA and monitored 3 days later for levels of productive infection. Results are gated on live, singlet CD3+CD8- cells. Numbers correspond to the percentage of infected cells in each sample. (B) t-SNE plots of uninfected and infected cells from Donor #1 shown in panel A, where the PBMC and endometrial specimens were run as separate t-SNEs. Infected cells reside in unique regions of the t-SNE as a result of viral-induced remodeling. Data are representative of 4 independent donors. (C) t-SNE plots of uninfected and infected cells from Donor #1 shown in panel A, where the PBMC and endometrial specimens were run within the same t-SNE. Although the same number of HIV-infected ETs are shown here as in panel B, these cells occupy less t-SNE space since much of the space is now taken up by cells from the PBMC sample. Corresponding data for the remaining 3 donors in panel A are presented in Figure 2—figure supplement 2. (D) Schematic defining Predicted Precursor (‘PRE’) cells. A population of T cells is either mock-treated (top) or exposed to HIV (bottom). These cells include ones that are not susceptible to infection (yellow and green) and those that are (blue and purple). After 3 days, infected cells (surrounded by a black ring) are identified as those expressing the HIV reporter HSA. These cells, however, are remodeled as represented by the conversion from their original blue and purple colors to the red and pink colors, respectively. By using PP-SLIDE to identify for every infected cell the phenotypically most similar cell in the uninfected culture (upward arrows), we identify the PRE cells which harbor the predicted phenotypes of the original cells targeted for HIV infection. These cells do not have the confounder of HIV-induced phenotypic changes, and can be compared to non-susceptible cells in the uninfected culture to identify unique features associated with cells targeted for infection. (E) t-SNE plots of uninfected and PRE cells demonstrate that the dominant population of T cells targeted for infection in both blood and endometrium are memory CD4+ T (Tm) cells. Tm cells (CD4+CD45RO+CD45RA-), naïve CD4+ T cells (CD4+CD45RO-CD45RA+, abbreviated Tn cells), memory CD8+ T cells (CD8+CD45RO+CD45RA-), and naïve CD8+ T cells (CD8+CD45RO-CD45RA+) were identified by manual gating and colored as indicated. Corresponding data for the remaining 3 donors in panel A are presented in Figure 2—figure supplement 2. (F) Comparison of the proportions of Tem (CD4+CD45RO+CD45RA-CCR7-CD62L-) and Tcm (CD4+CD45RO+CD45RA-CCR7+CD62L+) among uninfected Tm and PRE cells in PBMCs and ETs reveals preferential infection of Tem in both compartments. *p<0.05, **p<0.01, ***p<0.001 as assessed using the Student’s paired t test.
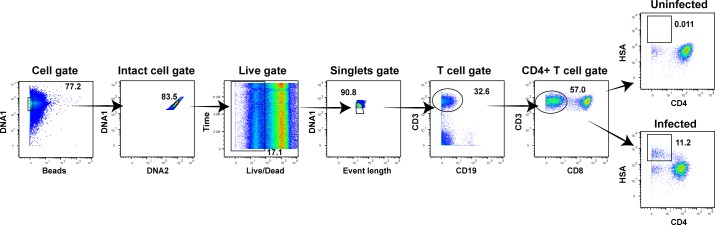
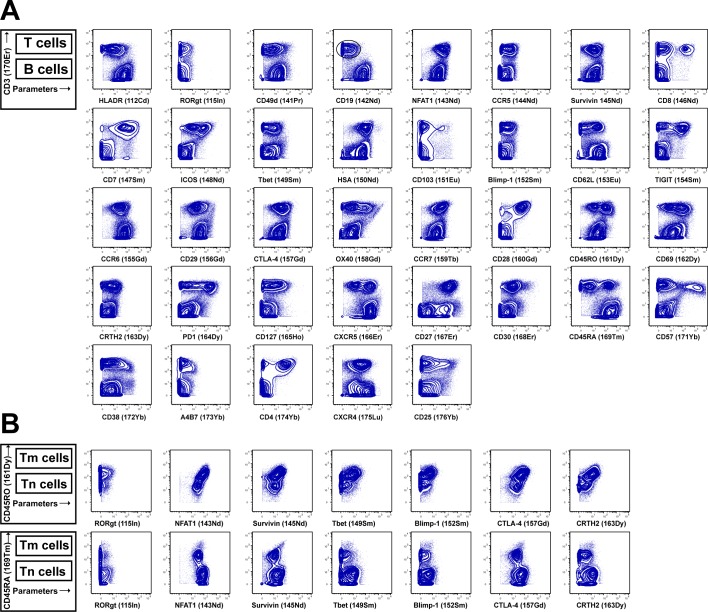
Figure 2—figure supplement 1. CyTOF gating strategy to identify uninfected and HIV-infected T cells.
Figure 2—figure supplement 2. Comparison of HIV-susceptible cells in unstimulated PBMCs and endometrium.
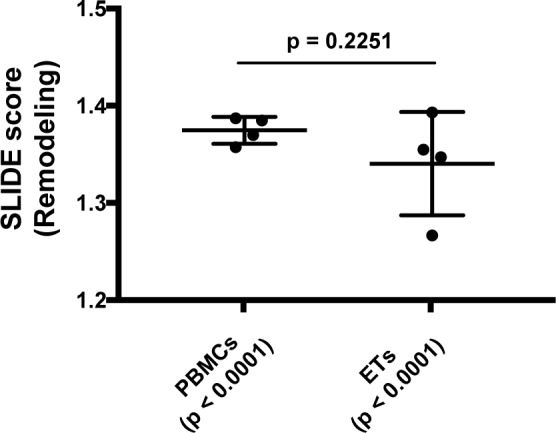
Figure 2—figure supplement 3. Quantitation of viral-induced remodeling of T cells from PBMCs and ETs.