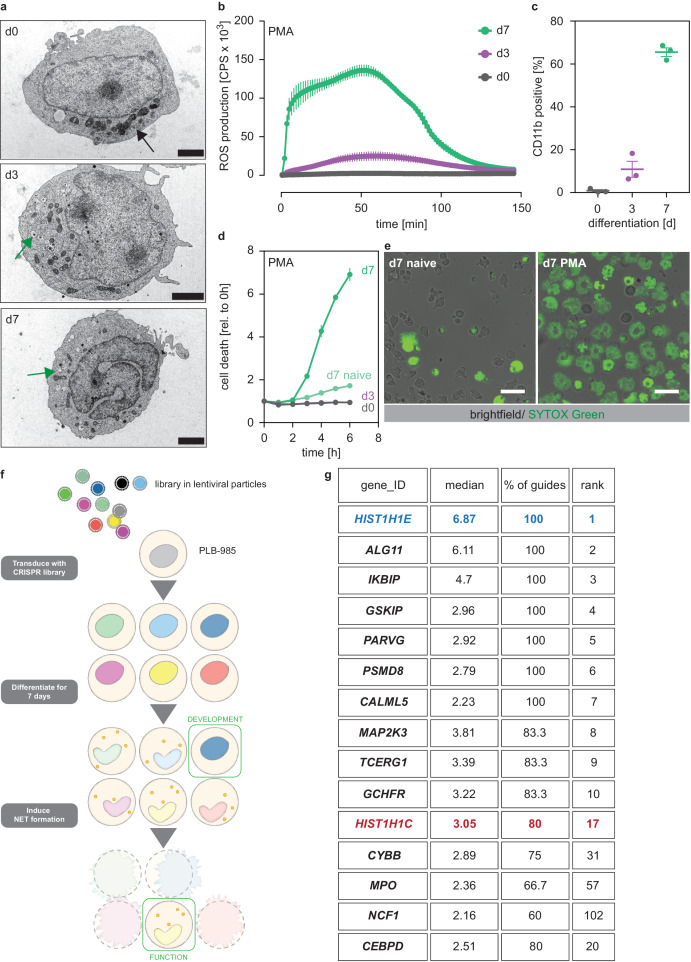
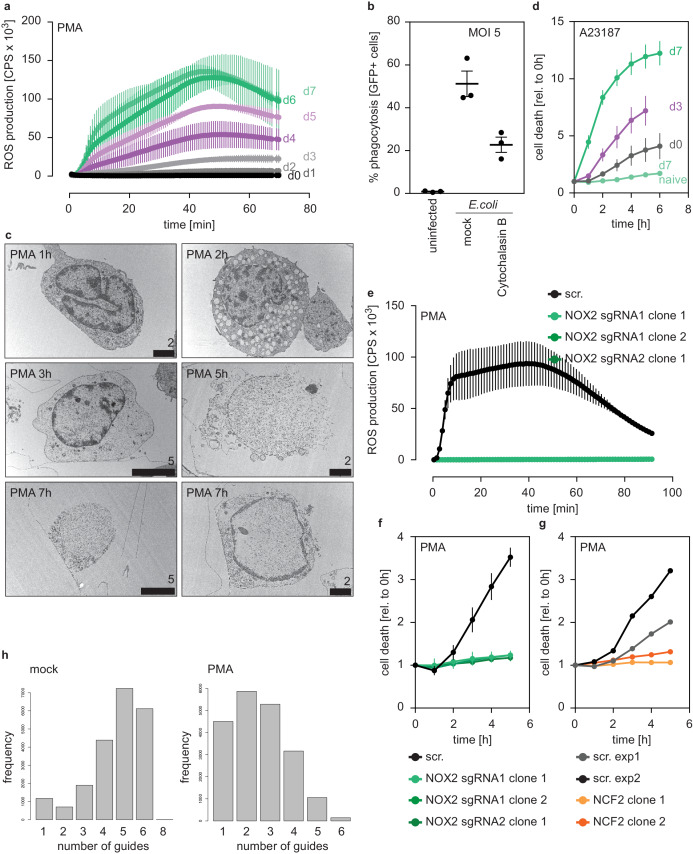
Figure 1. A genome-wide screen to identify genes required for PLB-985 cells differentiation and function.
(a–d) Characterization of PLB-985 differentiation and function at d0, d3 and 7 of differentiation. (a) Electron microscopy images of PLB-985 showing acquisition of granules (green arrows, the black arrow at d0 indicates mitochondria) and changes in nuclear morphology. Scale bars correspond to 5 μm. (b) ROS production of PLB-985 in response to 100 nM PMA, showing that only fully differentiated PLB-985 produced an oxidative burst. (c) Surface expression of CD11b, depicted is the percentage of CD11b positive cells out of all viable singlets. (d) PMA-induced cell death was measured over time after addition of the cell-impermeable DNA dye SYTOX Green and analysis of fluorescence indicating cell death. ‘d7 naïve’ indicates differentiated cells, which were not treated with PMA. (e) Representative images of differentiated PLB-985 (d7) after addition of SYTOX Green and treatment with or without PMA. Scale bars are 20 μm. (f) Outline of the CRISPR/Cas9 screen. Cells were transduced with a genome-wide CRISPR/Cas9 library (lentiviral particles, small circles), differentiated for 7 days, treated with PMA for 16 hr and survivors were sorted and sequenced to identify sgRNAs. (g) Top 10 of the screen, H1.2 (HIST1H1C) and indicated genes with known neutrophil functions, ranked first by % of overrepresented guides and then by median overrepresentation and mapped to a proteome list of human primary neutrophils. (b–d) Depicted are mean -/+ SEM of 3 independent experiments.