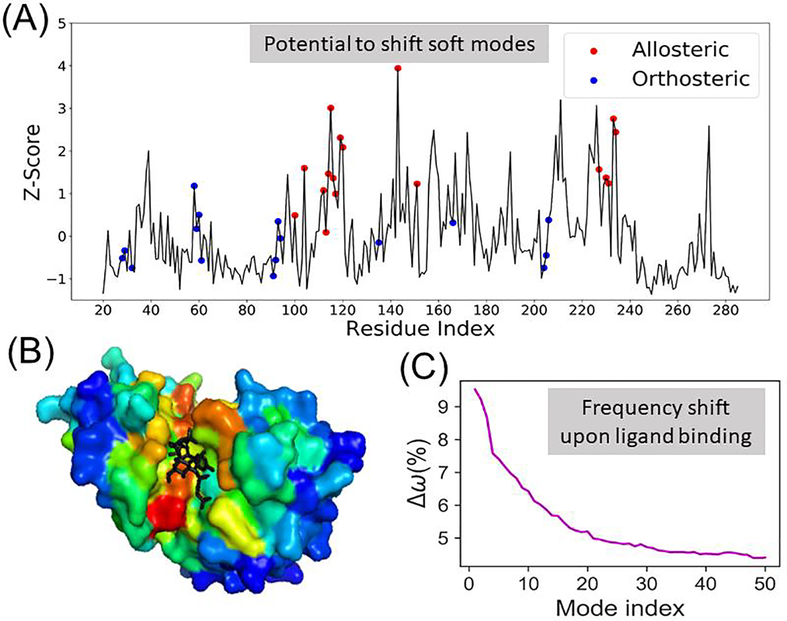
Fig 3. Potential of residues to induce a shift (increase) in the frequency of soft modes if targeted by a ligand.
Panel (A) illustrates the results obtained by scanning all residues in glutamate racemase (PDB id: 2JFN). The ordinate, z-score, gives a measure of the extent of frequency shifts in the global modes (averaged over ten softest modes), with peaks referring to those sites that would induce the highest shift, if targeted. Experimentally known binding/coordination sites for allosteric ligands (residues within 4.5 Å from allosteric ligands) are indicated by red circles, and those for orthosteric ligands, by blue circles. (B) Glutamate racemase in complex with an allosteric activator (UMA, shown in black sticks) and the orthosteric ligand (L-Glu, not seen from this perspective). The enzyme is color-coded by z-scores, red and blue, representing highest and lowest values, respectively. The allosteric ligand binding cavity is distinguished by its high potential to induce a frequency shift. (C) Results for a dataset of 315 protein complexes resolved in the presence of orthosteric and/or allosteric ligands. The curve displays the percent shift in the frequency of each of the 50 softest modes, computed by RESPEC, between the complex and the holo forms of the protein, averaged over all members of the dataset. The frequencies shift toward higher values, indicating a stiffening in the soft modes.