Figure 1.
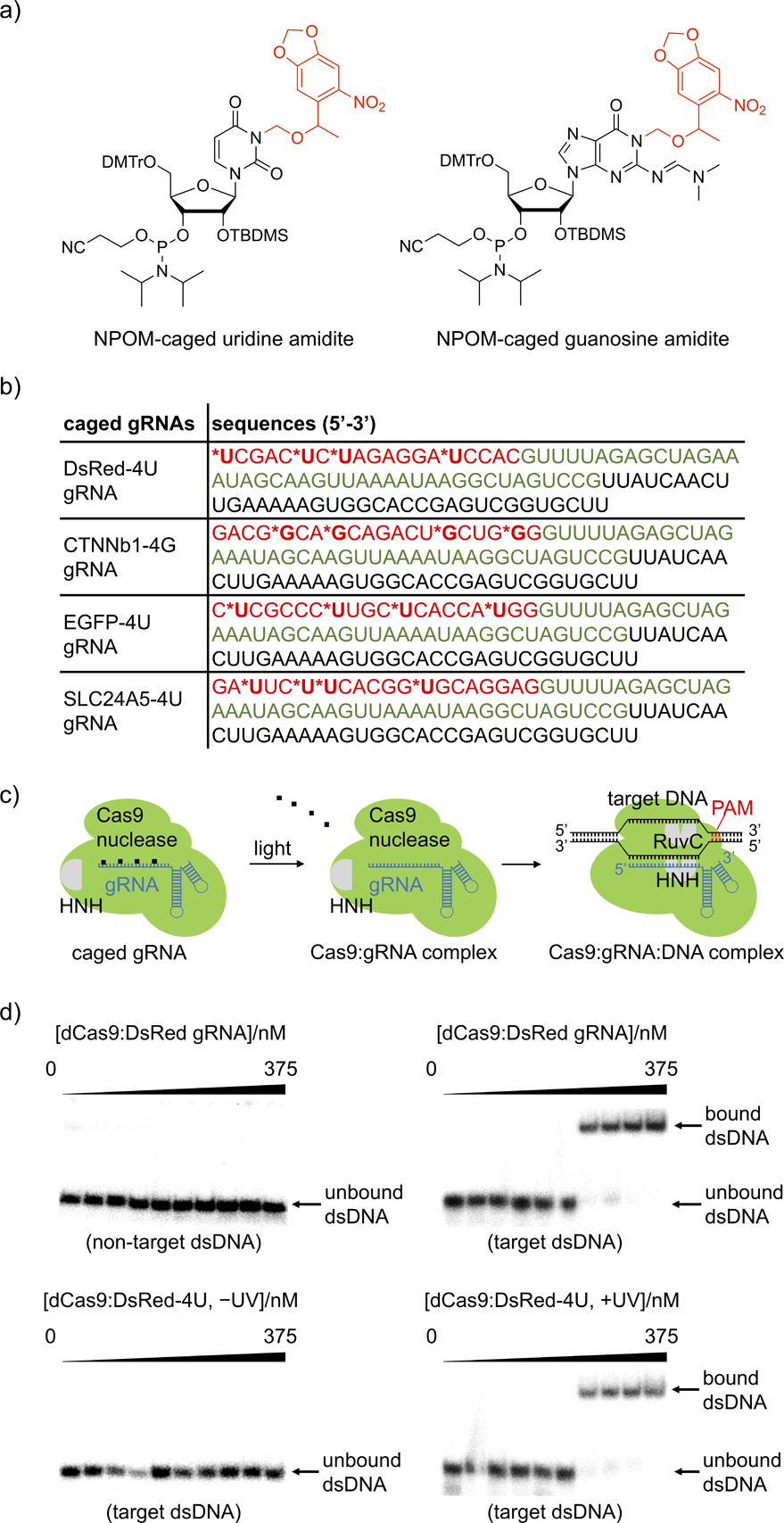
a) Structure of NPOM-protected uridine and guanosine amidites with photocleavable caging groups shown in red. b) Sequences of the photocaged gRNAs. The photocaged nucleotides are labelled by asterisks and the 20 nt base-pairing region of the gRNA is shown in red. The Cas9 binding region is shown in green and the S. pyogenes terminator region is shown in black. The corresponding non-caged gRNAs (DsRed, CTNNb1, EGFP, and SLC24A5) have the exact same sequences without the nucleobase caging groups. c) The NPOM-photocaging groups are designed to abolish RNP binding to the target dsDNA until they are photochemically cleaved, thereby generating an active Cas9:gRNA complex. d) Autoradiography of gel shift assays demonstrate that the photocaged gRNA effectively blocked the binding affinity of Cas9 to target 32P-labelled dsDNA and that binding is fully restored upon light activation, matching negative (non-target dsDNA) and positive (target dsDNA) controls. The exact concentrations of dCas9:gRNA used are 0, 0.0375, 0.1125, 0.375, 1.125, 3.75, 11.25, 37.5, 112.5, and 375 nM.
