Summary
Since December 2019, a novel coronavirus SARS-CoV-2 has emerged and rapidly spread throughout the world, resulting in a global public health emergency. The lack of vaccine and antivirals has brought an urgent need for an animal model. Human angiotensin-converting enzyme II (ACE2) has been identified as a functional receptor for SARS-CoV-2. In this study, we generated a mouse model expressing human ACE2 (hACE2) by using CRISPR/Cas9 knockin technology. In comparison with wild-type C57BL/6 mice, both young and aged hACE2 mice sustained high viral loads in lung, trachea, and brain upon intranasal infection. Although fatalities were not observed, interstitial pneumonia and elevated cytokines were seen in SARS-CoV-2 infected-aged hACE2 mice. Interestingly, intragastric inoculation of SARS-CoV-2 was seen to cause productive infection and lead to pulmonary pathological changes in hACE2 mice. Overall, this animal model described here provides a useful tool for studying SARS-CoV-2 transmission and pathogenesis and evaluating COVID-19 vaccines and therapeutics.
Keywords: SARS-CoV-2, mouse model, angiotensin-converting enzyme II, pathogenesis, hACE2-KI/NIFDC
Graphical Abstract
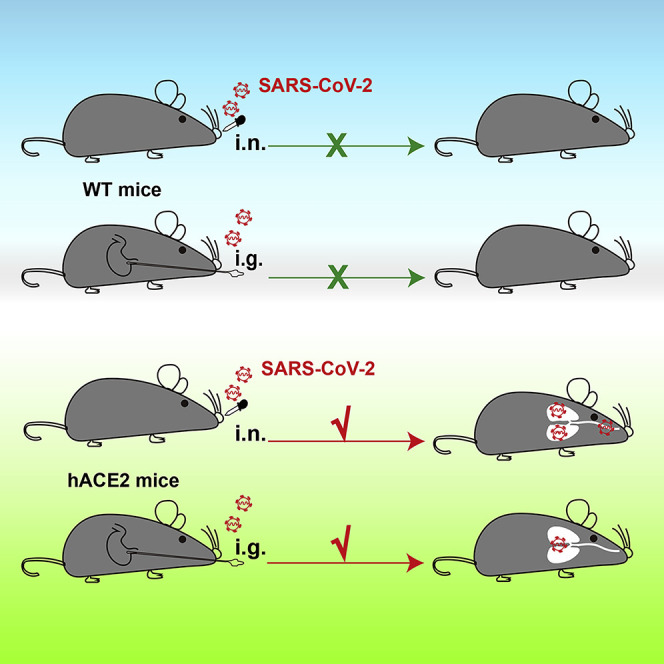
Highlights
-
•
Human ACE2 knockin mice were generated by using CRISPR/Cas9 technology
-
•
SARS-CoV-2 leads to robust replication in lung, trachea, and brain
-
•
SARS-CoV-2 causes interstitial pneumonia and elevated cytokine in aged hACE2 mice
-
•
High dose of SARS-CoV-2 can establish infection via intragastric route in hACE2 mice
The COVID-19 pandemic has brought an urgent need for small animal models. Here, Sun et al. established an ACE2 humanized mouse by CRISPR/Cas9 knockin technology. These hACE2 mice are susceptible to SARS-CoV-2 infection upon intranasal inoculation, and the resulting pulmonary infection and pathological changes resemble those observed in COVID-19 patients.
Introduction
Coronaviruses (CoVs) are a group of enveloped, positive-sensed, single-stranded RNA viruses within the subfamily Coronavirinae in the family Coronaviridae and the order Nidovirales. All coronaviruses can be divided into four genera: alphacoronaviruses, betacoronaviruses, gammacoronaviruses, and deltacoronaviruses. During the past decades, two highly transmissible and pathogenic CoVs, Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), have emerged in humans and caused global concerns. SARS-CoV was first recorded in Guandong, China, in December 2002 and rapidly spread to five continents (Peiris et al., 2003). The SARS-CoV outbreak ultimately caused 8,437 infections and 813 deaths worldwide, according to the World Health Organization (WHO). MERS-CoV first emerged in the Arabian Peninsula in 2012 (Zaki et al., 2012) and has caused 1,728 confirmed cases including 624 deaths in Middle Eastern countries, the United Kingdom, and South Korea, according to WHO. Both SARS-CoV and MERS-CoV likely originated from wild animals and are prone to cause lethal pneumonia in humans (Cui et al., 2019). There is currently no vaccine or specific antiviral drugs to prevent or treat their infections and transmission in humans.
In December 2019, a cluster of pneumonia patients with links to Huanan seafood market, and with potential animal contact, was reported in Wuhan, Hubei Province, China (Li et al., 2020, Zhou et al., 2020b, Zhu et al., 2020). Subsequently, the disease rapidly spread throughout the world to become a pandemic. The etiologic agent of the disease was then identified as a novel coronavirus by several independent groups (Wu et al., 2020, Zhou et al., 2020b) and has now been named as SARS-CoV-2 because of its close relationship to SARS-CoV. The disease was subsequently termed coronavirus disease 19 (COVID-19). The most common clinical manifestations of the patients are fever, cough, shortness of breath, and fatigue, and some patients have radiographic ground-glass lung changes and finally died from acute respiratory distress syndrome (ARDS) (Huang et al., 2020, Wang et al., 2020a).The WHO declared COVID-19 as a public health emergency of international concern on January 30, 2020.
Full genome-sequencing analysis has classified SARS-CoV-2 as a new member of the betacoronavirus genus, which includes SARS-CoV, MERS-CoV, and bat SARS-related coronaviruses (SARSr-CoV). SARS-CoV-2 has 82% nucleotide identity with SARS-CoV and 89% identify with a bat CoV, RaTG13, discovered in a cave of Yunnan, China, in 2013 (Lu et al., 2020, Zhou et al., 2020b). Like other CoVs, the genome RNA of SARS-CoV-2 encodes a non-structural replicase polyprotein and structural proteins, including spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins. The S protein of CoVs constitutes the spike on the virion surface and plays a vital role in viral binding, fusion, and entry as well as the induction of neutralizing antibody and T cell responses (Du et al., 2009). More importantly, several pioneering investigations have demonstrated that both SARS-CoV-2 and SARS-CoV utilize the same cellular receptor, human angiotensin-converting enzyme 2 (hACE2) to enter cells (Walls et al., 2020, Li et al., 2003, Zhou et al., 2020b). The crystal structure of the SARS-CoV-2 S protein receptor-binding domain (RBD) binding to hACE2 has been resolved, with an estimated 10- to 20-fold higher affinity toward hACE2 than SARS-CoV has (Lan et al., 2020, Shang et al., 2020). These findings pave the way for potent intervention strategies targeting RBD/hACE2 interactions. Exogenous hACE2 has been demonstrated to inhibit SARS-CoV-2 infection in cell cultures (Monteil et al., 2020, Ou et al., 2020), and several monoclonal antibodies targeting RBDs of SARS-CoV or SARS-CoV-2 are being tested with in vitro cultures (Walls et al., 2020). At present, no vaccine or antiviral drugs have been tested effectively in animal models.
Small animal models that reproduce the clinical course and pathology observed in COVID-19 patients are highly needed. In the present study, we generated a stable humanized ACE2 mice by using CRISPR/Cas9 knockin technology, replacing the endogenous mouse ACE2 (mACE2) with the human ACE2. Further characterization demonstrated that these humanized mice are highly susceptible to SARS-CoV-2 infection via intranasal inoculation, and the viral kinetics and pathological outcomes seen in the SARS-CoV-2-infected animals recapitulated the major finding from COVID-19 patients. Additionally, intragastric injection of SARS-CoV-2 could also establish infection and cause lung injury in the humanized ACE2 mice. This unique mouse model described here provides a useful tool for studying SARS-CoV-2 infection and transmission, as well as evaluating SARS-CoV-2 vaccines and therapeutics.
Results
An hACE2 Humanized Mouse Was Established by Using CRISPR/Cas9
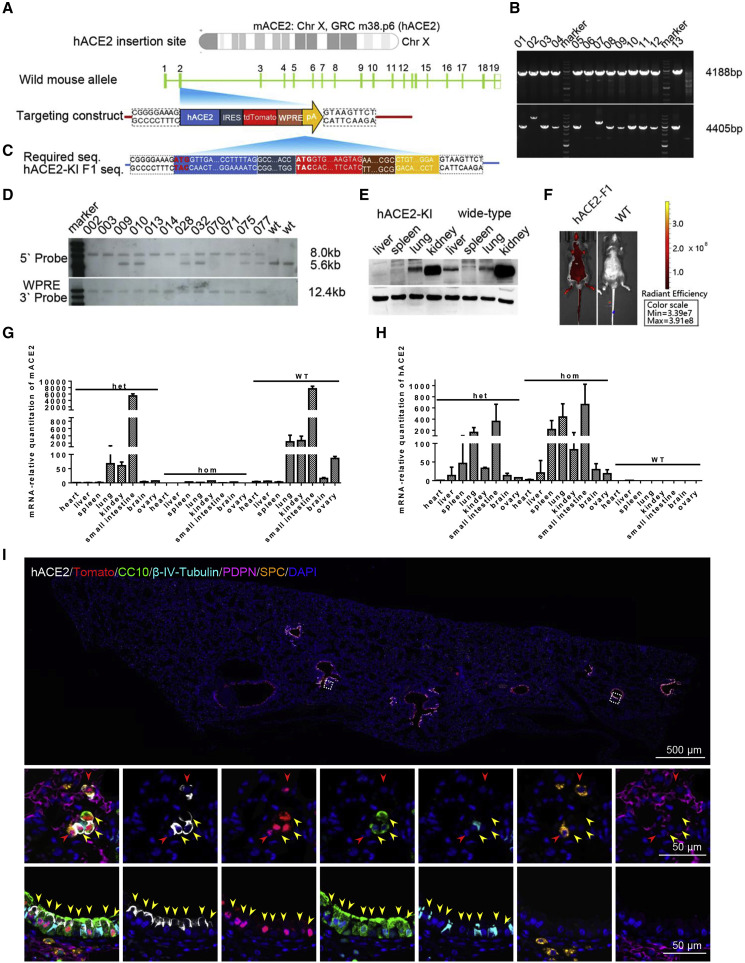
Previous studies have demonstrated that transgenic mice expressing human ACE2 became highly susceptible to SARS-CoV infection (McCray et al., 2007, Menachery et al., 2016, Netland et al., 2008, Tseng et al., 2007, Yang et al., 2007). Here, we aimed to establish a humanized ACE2 mouse by using CRISPR/Cas9 knockin technology. As shown in Figure 1 A, the full cDNAs of hACE2 were inserted into Exon 2 (Figure 1A), the first coding exon, of the mAce2 gene located in GRC m38.p6 sites on chromosome X. This disrupted the mAce2 gene and terminated its expression. The tdTomato gene was inserted downstream of hAce2 with an internal ribosome entry site (IRES), allowing co-expression of hACE2 and tdTomato. A woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) and poly (A) sequence were added to enhance mRNA stability and translation efficiency. This targeting strategy allowed an intrinsic expression profile of hACE2 under control of mAce2 promoter. Along with subgenomic RNA (sgRNA) and Cas9 mRNA, the targeting construct was injected into 370 zygotes of C57BL/6 mice. Successful insertion in 10 of 46 offspring (21.74%) was confirmed by PCR screen (Figure 1B); then three founders were backcrossed with C57BL/6 mice, and 37 F1+ offspring were screened. The targeting results of 12 positive F1 mice were further confirmed by sequencing (Figure 1C) and Southern blotting (Figure 1D). As expected, no animals had random insertion as demonstrated by the WPRE internal probe (Figure 1D). Importantly, western blotting results showed that hACE2 expression was strong in lung and kidney but very weak in spleen and liver, which displayed a similar expression pattern of mACE2 protein in wild-type (WT) animals (Figure 1E). More importantly, the mAce2 gene was not detected by PCR in homozygous hACE2 mice and decreased largely in heterozygous mice (Figure 1F), whereas significant hACE2 expression was detected in lung, small intestine, spleen, and kidney from both hACE2 heterozygous and homozygous mice but not in the WT mice (Figure 1G). Profiting from the co-expression design, the rough global expression patterns of hACE2 were examined visually via bioluminescent imaging in the hACE2 mice (Figure 1H). Furthermore, immunofluorescence staining of the lung sections from the homozygous mice showed that hACE2, as well as tdTomato, were predominantly expressed in the CC10+ Clara cells along the airway, as well as a small population of surfactant protein C positive (SPC+) alveolar type II cells (Figure 1I). Thus, we established a homozygous mouse model that stably expresses hACE2 under the control of mACE2 promoter, which were termed hACE2-KI/NIFDC mice (abbreviated to hACE2 mice) accordingly.
Figure 1.
Construction and Characterization of hACE2 Humanized Mouse
(A) The hACE2 gene were inserted into exon2, the first coding exon, of mouse ACE2 locating in chromosome X GRC m38.p6.
(B) The mice produced both 4,188 bp and 4,405 bp PCR amplicons that were identified as founder. Ten representative positive PCR results were shown.
(C) F1+ mice were sequenced to confirm the right insertion. The head and tail bases of each part were aligned with expected one.
(D) Two endonucleases, NcoI and StuI, were used to digest DNA for identifying the right insertion and no random recombinant. The expected sizes of WT and gene-targeted bands were 5.6 kb and 8.0 kb, respectively. Heterozygous female mice had both bands, whereas homozygous male mice had only one 8.0 kb band. All the mice detected with WPRE probe had only one 12.4 kb band, indicating no random insertion.
(E) western blotting results of tissue distribution of hACE2 in humanized mice and WT mice. The primary antibody used here cross-reacted with both mACE2 and hACE2.
(F and G) The expression pattern of mACE2 (F) and hACE2 (G) in hACE2 heterozygous and homozygous and WT mice (n = 3) detected by real-time qPCR.
(H) Bioluminescence imaging (BLI) of homozygous mice and WT mice.
(I) Immunofluorescence staining analysis for the hAEC2-expressed cell types in lung of hACE2 mice. Paraffin lung sections were stained for hACE2 (white), tdTomato (red) CC10 (green), β-IV-Tubulin (cyan), PDPN (magenta), and SPC (gold). Two white frames were magnified below. Solid red and yellow arrows indicated the ACE2+/tdTomato+/SPC+ cells and ACE2+/tdTomato+/CC10+ cells, respectively.
The hACE2 Mice Are Susceptible to Intranasal Infection of SARS-CoV-2
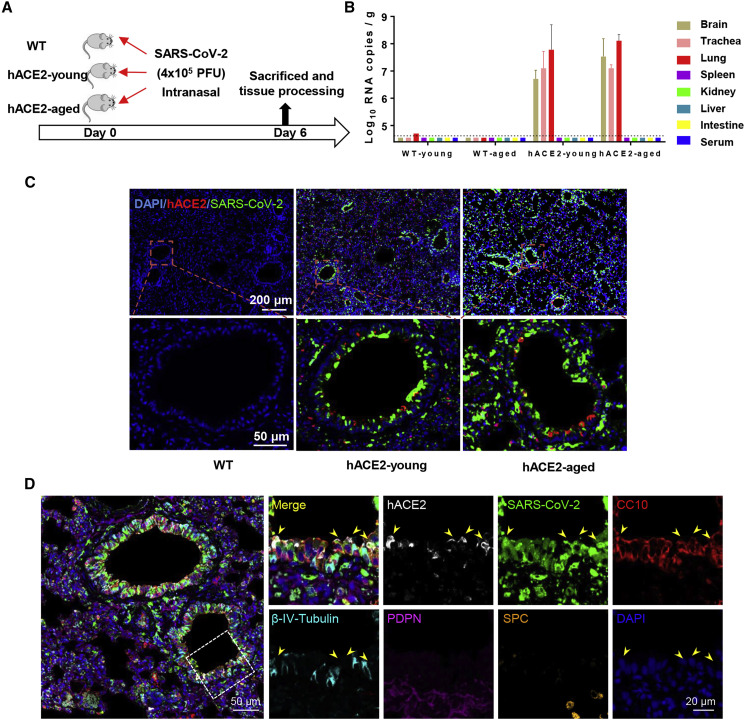
To further characterize the susceptibility of hACE2 mice to SARS-CoV-2 infection, groups of young (4.5-week-old) and aged (30-week-old) hACE2 mice were intranasally infected with 4 × 105 plaque-forming units (PFU) of SARS-CoV-2, and WT C57BL/6 mice that received the same dose of viral challenge were set as control (Figure 2 A). All animals were monitored daily for clinical symptoms and body-weight changes and were sacrificed on day 6 post-infection (dpi). None of the inoculated animals displayed obvious clinical symptoms, and only the aged hACE2 mice lost 10% of their weight at 3 dpi and then recovered (Figure S1). As shown in Figure 2B, robust viral RNA replication was seen in lung, trachea, and brain tissues from both young and aged hACE2 mice, whereas no detectable viral RNA was seen in spleen, kidney, liver, intestine, and serum. Especially, infectious SARS-CoV-2 were successfully rescued from lung samples by using plaque assay. As expected, no viral RNAs were detected in any tested tissues or serum from the WT C57BL/6 mice (Figure 2). Interestingly, we also detected high levels of viral RNAs (2.9 × 105 copies/g) in feces of aged hACE2 mice. Further immunostaining of mouse lung section showed that robust viral S protein was detected along the airway in both young and aged hACE2 mice, and bare viral protein was detected in the SARS-CoV-2-inoculated WT mice (Figure 2C). To confirm the major target cells of SARS-CoV-2 in our model, we further stained the lung section from the infected hACE2 mice with different cell markers, including Clara cells (marked by Clara cell 10 kDa protein/CC10), ciliated cells (marked by β-IV-tubulin), alveolar type 1 cells (AT1, marked by podoplanin/PDPN), and alveolar type 2 cells (AT2, marked by surfactant protein C/SPC), as well as viral S protein and hACE2. As shown in Figure 2D, SARS-CoV-2 S protein predominantly colocalized with CC10 and hACE2, indicating that CC10+ Clara cells are the major target cells of SARS-CoV-2 along the airway in our model. Additionally, immunostaining of brain sections from the infected hACE2 mice showed that robust viral S protein expression could also be detected in neuron, astrocyte, and microglial cells (Figure S2). Together, these results indicate that the hACE2 mice are highly susceptible to intranasal infection of SARS-CoV-2 in comparison with WT mice, with sustained robust viral RNA replication in lung Clara cells as well as in trachea and brain.
Figure 2.
Intranasal Infection of SARS-CoV-2 in hACE2 Mice
(A) WT C57BL/6 mice, young (4.5-week-old), and aged (30-week-old) hACE2 mice (n = 3) were intranasally infected with 4 × 105 PFU of SARS-CoV-2, and all mice were sacrificed on 6 dpi for tissue collection.
(B) Tissue distribution of SARS-CoV-2 viral RNAs. Each tissue and serum were processed with viral RNA copies analysis by real-time qPCR. Dotted lines denote the detection limit.
(C) Immunofluorescence staining of mouse lung paraffin sections for SARS-CoV-2 S protein (green), ACE2 (red), and DAPI (blue).
(D) Immunofluorescence staining analysis for the infected cells in lung. Cell markers for hACE2 (white), CC10 (red), β-IV-Tubulin (cyan), PDPN (magenta), and SPC (gold). White frame was magnified on the right. Solid yellow arrows indicate the SARS-CoV-2+/hACE2+/CC10+ cells.
hACE2 Mice Developed Interstitial Pneumonia upon SARS-CoV-2 Infection
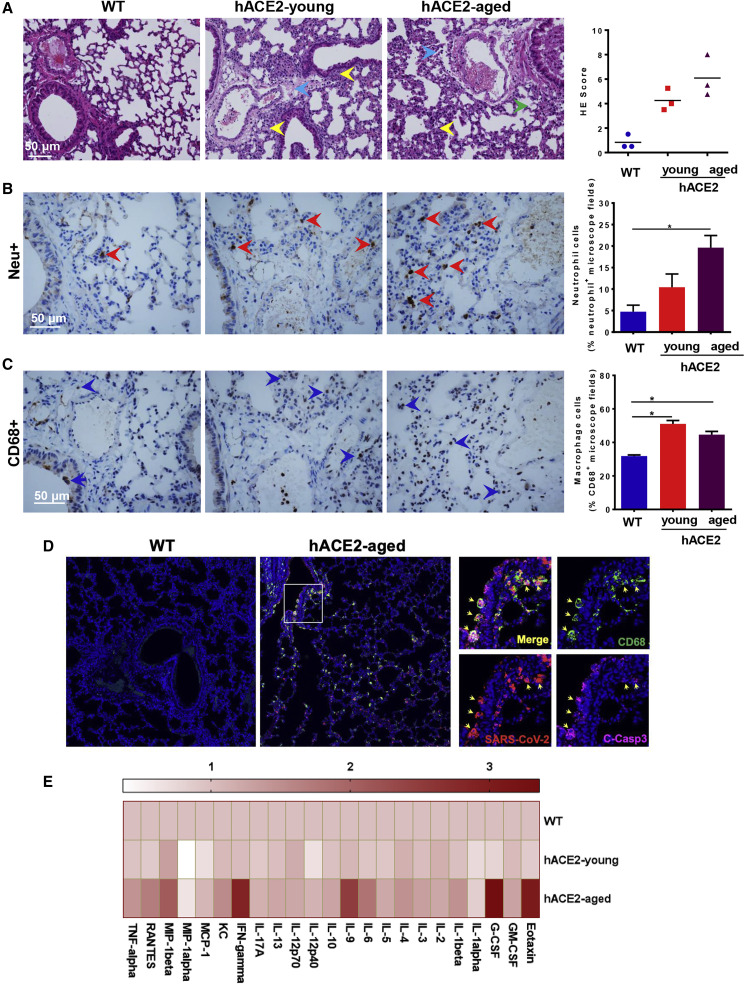
To check whether the SARS-CoV-2-infected hACE2 mice produced similar pathological features as those observed in COVID-19 patients, histopathological examination, immunohistochemistry (IHC), and immunofluorescence staining were performed in lung sections from SARS-CoV-2-infected animals. In comparison with SARS-CoV-2-infected WT mice, H&E staining showed that both young and aged hACE2 mice developed interstitial pneumonia characterized with inflammatory cell infiltration, alveolar septal thickening, and distinctive vascular system injury. More lesions of alveolar epithelial cells and focal hemorrhage were observed in the aged mice (Figure 3 A). In comparison with WT mice, IHC staining analysis showed SARS-CoV-2 infection induced more neutrophil (Neu+) and macrophage (CD68+) infiltration in the aged hACE2 mice (Figures 3B and 3C). Additionally, immunofluorescence co-staining showed SARS-CoV-2 directly infected CD68+ macrophages in lung, resulting in significant apoptosis (C-Casp3+) in the aged hACE2 mice (Figure 3D). Most importantly, Luminex cytokine analysis showed that SARS-CoV-2 infection led to elevated cytokine production including Eotaxin, G-CSF, IFN-γ, IL-9, and MIP-1β in aged hACE2 mice, but it had a weaker response in young mice (Figure 3E; Table S1). Thus, the hACE2 mice not only sustained viral replication but also developed pulmonary pathology upon SARS-CoV-2 infection.
Figure 3.
Pathological Changes and Inflammatory Response in SARS-CoV-2-Infected hACE2 Mice
(A) H&E staining analysis showing inflammatory cell infiltration (yellow arrow), alveolar septal thickening, focal hemorrhage (green arrow), and distinctive vascular system injury (blue arrow) in hACE2 mice. Right panel, semiquantitative analysis of the H&E-stained lung sections.
(B and C) IHC staining analysis for neutrophils (Neu+) and macrophages (CD68+) in SARS-CoV-2 infected hACE2 mice. Right panel, semiquantitative analysis of the neutrophil and macrophage counts. Statistical significance was analyzed by unpaired Student’s t tests. ∗p < 0.05.
(D) Immunofluorescence staining analysis for paraffin lung sections with markers for CD68 (green), cleaved-caspase 3 (C-Casp3, magenta), and SARS-CoV-2 S protein (red). White frame was magnified on the right. Solid yellow arrows indicated the SARS-CoV-2+/CD68+/C-Casp3+ cells.
(E) Cytokine production in serum of SARS-CoV-2-infected mice.
SARS-CoV-2 Established Productive Infection via Intragastric Route in hACE2 Mice
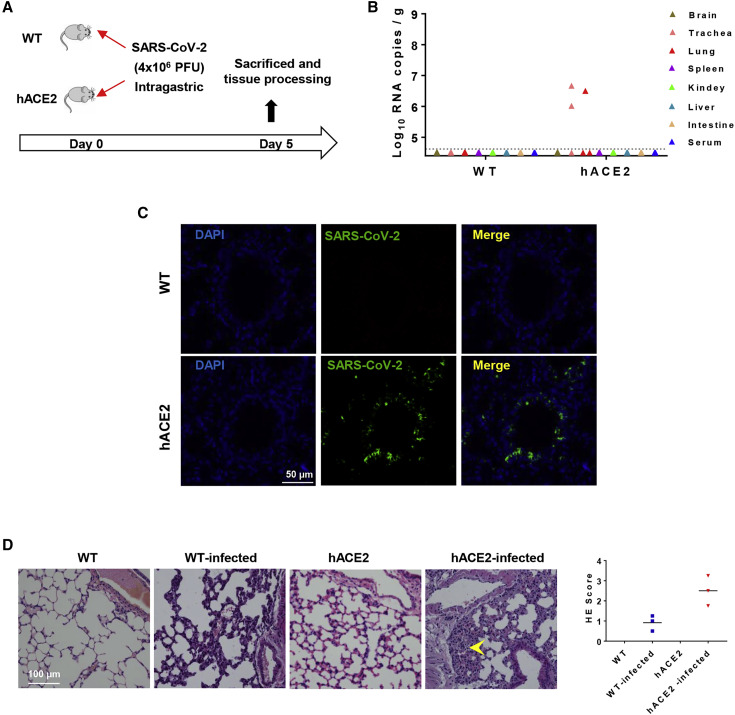
SARS-CoV-2 mainly transmits via droplets and close contact among humans. However, some patients with COVID-19 infection had gastrointestinal symptoms such as diarrhea, abdominal pain, and vomiting (Lin et al., 2020, Wang et al., 2020a), and viral RNAs have been detected in feces of COVID-19 patients (Cai et al., 2020, Xu et al., 2020), highlighting the possibility of fecal-oral transmission of SARS-CoV-2 (Yeo et al., 2020). Here, we sought to investigate whether SARS-CoV-2 could establish productive infection via intragastric route in hACE2 mice. After intragastric administration of SARS-CoV-2, all animals were subjected to daily monitoring and tissue processing on 5 dpi (Figure 4 A). Although no clinical signs were seen in the three inoculated hACE2 mice, high levels of viral RNA were detected in trachea (2.9 × 106 copies/g) and lung (3.2 × 106 copies/g) of two animals (Figure 4B), which was comparable to that in animals infected via intranasal route. Importantly, the expression of SARS-CoV-2 S protein was also detected along the lung airway in the infected hACE2 mice (Figure 4C). In addition, interstitial inflammation was observed with alveolar septal thickening in the intagastric SARS-CoV-2-infected hACE2 mice (Figure 4D). Overall, our results demonstrated that intragastric administration of SARS-CoV-2 could also establish productive infection and lead to pulmonary pathological changes in hACE2 mice.
Figure 4.
Intragastric Infection with SARS-CoV-2 in hACE2 Mice
(A) WT C57BL/6 mice and hACE2 mice (n = 3) at 4.5 weeks old were infected with 4 × 106 PFU of SARS-CoV-2 through intragastric administration, and all mice were sacrificed on 5 dpi for main tissues and serum collection.
(B) Tissue distribution of SARS-CoV-2 viral RNAs. Each tissue and serum were collected and viral RNA copies analysis by real-time qPCR.
(C) Immunofluorescence staining of mouse lung sections for SARS-CoV-2 S protein (green) and DAPI (blue).
(D) Histopathological analysis of mouse lung sections. Alveolar septal thickening was indicated with yellow arrow. Right panel, semiquantitative analysis of the H&E-stained lung sections.
Discussion
In the present study, we generated a stable humanized mouse strain in which hACE2 expression is under the control of IRES in fusion with the tdTomato reporter gene. Further characterization demonstrated that the expression of mACE2 was completely replaced by hACE2 in the homozygous mice with peak hACE2 expression in lung and small intestine (Figure 1). Previously, several hACE2 transgenic mice have been generated with different strategies and used for modeling SARS-CoV infection (McCray et al., 2007, Tseng et al., 2007). Generally, the pulmonary pathology caused by SARS-CoV in transgenic mice correlated with the level of hACE2 expression in lung. Thus, such transgenic mouse has been explored to mimic SARS-CoV-2 infection (Bao et al., 2020). When compared with such transgenic mouse model, our hACE2 mouse model has several advantages. First, the hAce2 gene was inserted at GRC m38.p6 sites on chromosome X to replace mAce2, and there is no endogenous mACE2 expression in homozygous hACE2 mice. Because transgene insertion occurs randomly, the integrity of the landing genes would probably be affected. Second, the tissue distribution of hACE2 in our mouse model matches the clinical findings from COVID-19 patients, and high levels of hACE2 expression were detected in lung (Figure 1E). Interestingly, recent analysis based on single-cell sequencing results from human tissues showed that kidney, heart, esophagus, bladder, and ileum also represent tissues of high abundance (Qi et al., 2020, Zou et al., 2020), whereas liver, spleen, small intestine, ovary, and brain are the hACE2-abundant tissues in our model (Figure 1H); this inconsistency between human and mice deserves further investigation. Third, the introduction of tdTomato gene allows real-time monitoring of hACE2 expression. All these ideal physiological features, stable inheritances, and phenotypes of hACE2 mice prompted us to test their application in reproducing SARS-CoV-2 infection.
Not surprisingly, our challenge experiment of SARS-CoV-2 clearly demonstrated that these hACE2 mice are highly susceptible to SARS-CoV-2 infection via intranasal inoculation, and 4 × 105 PFU of SARS-CoV-2 readily led to robust viral replication in respiratory tracts (lung and trachea) as well as in brain. The peaked viral RNA loads in lung reached 108 copies/g, which is much higher than that in hACE2 transgenic mice (Bao et al., 2020). The presence of viral RNAs in brain was somewhat unexpected, because few COVID-19 patients developed neurological systems (Chen et al., 2020). Previous findings from a SARS-CoV-infected transgenic model also confirmed viral tropism to brain (McCray et al., 2007), which is consistent with our findings. The underlying mechanism through which SARS-CoV or SARS-CoV-2 invade the brain and the correlation to disease severity remain to be determined. Further characterization demonstrated that CC10+ Clara cells along the airway are the major target cells of SARS-CoV-2 (Figure 2D) in our model. Specific protease in Clara cells, like tryptase Clara, might contribute to the efficient cleavage of S protein to enhance viral entry into cells. Additionally, CD68+ macrophages in pulmonary alveolus are also infected with SARS-CoV-2, resulting in severe cell apoptosis (Figure 3D). Interestingly, a significant population of cells that do not express hACE2 were also infected with SARS-CoV-2 (Figure 2D). How SARS-CoV-2 invades these cells also deserves further investigation. Overall, our result provides the first line of evidence showing the specific major target cells of SARS-CoV-2 in mouse lung.
Most importantly, all hACE2 mice intranasally challenged with SARS-CoV-2 developed interstitial pneumonia characterized with inflammatory cell infiltration, alveolar septal thickening, and distinctive vascular system injury, which recapitulated the clinical features in most COVID-19 patients. Early epidemiological findings have suggested that advanced age resulted in increased mortality of COVID-19 patients (Chen et al., 2020, Yang et al., 2020, Zhou et al., 2020a), and our results from the hACE2 model reflect this finding well. The aged hACE2 mice (30 weeks old) lost weight upon SARS-CoV-2 infection, whereas young hACE2 mice (4.5 weeks old) as well as WT mice maintained their body weight increase during the observation period (Figure S1). Remarkably, although both young and aged mice sustained similar viral replication in lung, more severe pathological changes as well as increased cytokine response were seen in the aged hACE2 mice (Figure 3). These unique features in aged hACE2 mice resembling clinical findings (Wang et al., 2020b) highlight the potential application in different settings. It should be noted that no clinical symptoms or mortality was observed in our model, and other animal models that can recapitulate severe or lethal disease of COVID-19 should be warranted in the future.
There are several lines of evidence that support potential fecal-oral transmission of SARS-CoV-2. First, gastrointestinal symptoms were seen in many COVID-19 patients (Jin et al., 2020, Lin et al., 2020), and viral shedding in feces have been well reported. Second, ACE2 was found to be highly expressed in gastrointestinal epithelial cells, and in vitro studies also indicated SARS-CoV-2 can infect multiple cells from gastrointestinal tracts. Third, experience from SARS and MERS supported that human CoVs maintained their viability in environmental conditions essential for fecal-oral transmission (Chan et al., 2011, van Doremalen et al., 2013). Recent data from SARS-CoV-2 revealed similar stability to SARS-CoV, suggesting similar transmission routes are plausible (van Doremalen et al., 2020). Our results from hACE2 mouse model provide evidence supporting this hypothesis. Upon intranasal infection, viral RNA shading was also confirmed in feces from the hACE2 mice (data not shown). Due to facility limitation, we were not able to collect and quantitate feces samples from each individual animal. Importantly, SARS-CoV-2 given intragastrically caused productive infection in the respiratory tracts of hACE2 mice, as demonstrated by the presence of high level of viral RNAs and active viral protein expression (Figures 4B and 4C). Our preliminary results show, however, that intragastric infection of SARS-CoV is not as efficient as intranasal infection: the challenge dose of intragastric model is 10-fold higher than the intranasal model; however, pulmonary infection was only detected in one of three animals tested (Figure 4B); the viral RNA loads and protein expression were also lower than that in the intranasal model. Finally, whether intragastric infection of SARS-CoV-2 is actually occurring in clinical setting in humans remains to be confirmed. The underlying mechanism through which SARS-CoV-2 survives the gastrointestinal environment and finally invades the respiratory tracts in our mouse model warrants further investigation.
In summary, our results show that hACE2 mice are susceptible to intranasal and intragastric infection of SARS-CoV-2, and robust viral replication was seen in the pulmonary Clara cells and macrophages. Especially, the pathological changes observed in the aged hACE2 mice more closely resemble those observed in COVID-19 patients. Thus, the use of hACE2 mice described in our manuscript provides a small animal model for studying the transmission and pathogenesis of SARS-CoV-2 and for the understanding of unexpected clinical manifestations of SARS-CoV-2 infection in humans. This model will also be valuable for testing vaccines and therapeutics to combat SARS-CoV-2.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ACE2 (immunostaining) | Abcam | Cat#ab108209; RRID: AB_10862654 |
| SARS-CoV Spike | Sinobiological | Cat#40150-T52 |
| β-IV-tubulin | Abcam | Cat#ab179504 |
| CC10 | Millipore | Cat#07-623; RRID: AB_310759 |
| Podoplanin | Sinobiological | Cat#50256-R066 |
| SPC | Abcam | Cat#ab211326 |
| CD68 | Abcam | Cat#ab125212 |
| Cleaved Caspase-3 | CST | Cat#9661; RRID: AB_2341188 |
| Neutrophil Marker | Santa Cruz Biotechnology | Cat#sc-71674; RRID: AB_2167794 |
| GAPDH | Abcam | Cat#ab9485; RRID: AB_307275 |
| horseradish peroxidase (HRP)-conjugated anti-mouse IgG | Abcam | Cat#ab6728; RRID: AB_955440 |
| horseradish peroxidase (HRP)-conjugated anti-rabbit IgG | Abcam | Cat#ab205718; RRID: AB_2819160 |
| ACE2 (Western Blotting) | Huabio | Cat#ET1611-58; RRID: AB_11158910 |
| Virus Strains | ||
| SARS-CoV-2 BetaCoV/Wuhan/AMMS01/2020 |
This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DMEM | GIBCO | Cat#11995-065 |
| Fetal Bovine Serum | PAN Biotech | Cat#P30-3306 |
| Penecillin Streptomycin | GIBCO | Cat#15140-122 |
| HEPES | GIBCO | Cat#15630-080 |
| RNAlater | Invitrogen | Cat#AM7020 |
| retrieval/elution buffer | Abcracker | Cat#ABCFR5L |
| Streptavidin-peroxidase conjugate | Zhongshan Biotechnology | Cat#SP-9000 |
| low-melting point agarose | Promega | Cat#V2111 |
| Pierce ECL Western Blotting Substrate | Thermo | Cat#32106 |
| pelltobarbitalum natricum | Sigma | Cat#P3761 |
| NcoI | Thermo | Cat#FD0595 |
| StuI | Thermo | Cat#ER0421 |
| Critical Commercial Assays | ||
| RNeasy Mini kit | QIAGEN | Cat#74106 |
| One Step PrimeScript RT-PCR Kit | TaKaRa | Cat#RR064A |
| Bio-Plex Pro Mouse Cytokine Grp I Panel 23-Plex | BIO-RAD | Cat#M60009RDPD |
| Reverse transcription (RT)-PCR kit | TaKaRa | Cat#RR096A |
| NEON 7-color Allround Discovery Kit | Histova Biotechnology | Cat#NEFP750 |
| Experimental Models: Cell Line | ||
| Vero Cells | ATCC | Cat#CCL-81 |
| Experimental Models: Animals | ||
| C57BL/6 | Institute for Laboratory Animal Resources, NIFDC, China | N/A |
| hACE2-KI/NIFDC | This paper | N/A |
| Oligonucleotides | ||
| sgRNA gaaagatgtccagctcctcctgg |
This paper | N/A |
| Primers for genotyping F0 offspring, see Table S2 | This paper | N/A |
| Primers for RT-qPCR quantification of ACE2 expression level, see Table S2 | This paper | N/A |
| Primers and probes for RT-qPCR quantification of SARS-CoV-2 RNA, see Table S2 | This paper | N/A |
| Software and Algorithms | ||
| GraphPad Prism 7.0 | Graphpad | https://www.graphpad.com/ |
| Luminex PONENT | Thermo Fisher | https://www.luminexcorp.com/xponent/ |
| Other | ||
| IVIS-Lumina II imaging system | Xenogen | https://health.usf.edu/ |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, You-Chun Wang (wangyc@nifdc.org.cn).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
This study did not generate any unique datasets or code.
Experimental Model and Subject Details
Experimental Animals
Groups of 4.5 weeks and 30 weeks old hACE2 and wild-type C57BL/6 female mice for SARS-CoV-2 infection were maintained in animal biosafety level 3 (ABSL3) containment laboratory of AMMS at 23°C. All animals were allowed free access to water and diet and provided with a 12 h light/dark cycle. All animal experiments were approved by the Experimental Animal Committee of NIFDC (approval number: 2018-B-002) and Laboratory Animal Center, AMMS (approval number: IACUC-DWZX-2020-001).
Cell Lines
Vero Cells for SARS-CoV-2 propagation and titeration were maintained in DMEM medium (GIBCO) supplemented with 2% FBS (PAN Biotech), 1% Penecillin Streptomycin (GIBCO) and 1% HEPES (GIBCO) at 37°C.
SARS-CoV-2 Propagation and Titeration
SARS-CoV-2 strain BetaCoV/wuhan/AMMS01/2020 was originally isolated by CFQ’s lab, AMMS, from a patient returning from Wuhan. The virus was amplified on Vero cells and titerated by standard plaque forming assay. Briefly, Vero cells in 12-well plates were infected with a 10-fold serial dilution of viruses. The plates were incubated at 37°C for 1 h and cells were overlaid with 1% low-melting point agarose (Promega) in DMEM containing 2% FBS. After further incubation at 37°C for 2 days, the cells were fixed with 4% formaldehyde and stained with 0.2% crystal violet to visualize the plaques. All experiments involving infectious SARS-CoV-2 were performed in biosafety level 3 (BSL3) containment laboratory in AMMS.
Method Details
Generation of hACE2 Mice
To construct the targeting vector, cDNAs encoding hACE2 linked to the red fluorescent protein tdTomato by an IRES sequence, flowed with WPRE and ployA, was inserted into the first coding exon of mACE2 by homologous recombination. The expression of inserted hACE2 and tdTomato gene were driven by the mouse mACE2 promoter. The specific targeting sgRNA (5′-gaaagatgtccagctcctcctgg-3′) and Cas9 mRNA were microinjected into C57BL/6 zygotes, which were subsequently implanted into pseudo-pregnant mice. F0 offspring were genotyped using two pairs of primers L-GT-F1, L-GT-R1 and R-GT-F1, R-GT-R1 (Table S2). The expected size of amplicons was 4188 bp and 4405 bp, respectively. Genotype-positive founders were backcrossed with C57BL/6 mice to produce generation F1. DNA samples of F1-positive mice were subjected to Southern blotting to confirm off-targeting (by WPRE 3′ probe) and correct insertion (by 5′ probe). Two endonucleases, NcoI (Thermo) and StuI (Thermo), were used to digest DNA for identifying the right insertion and no random recombinant.
RNA Extraction and Real-Time Quantitative PCR
To confirm the expression of hACE2 and mACE2 in humanized mice, tissues from 4.5 weeks old and 30 weeks old mice were dissected, immediately immersed in RNAlater (Invitrogen) stabilization reagent and stored at −80°C. Total RNA was extracted from individual tissues using TRIzol and quantified using a spectrophotometer. Real-time quantitative PCR (RT-qPCR) was performed using a reverse transcription (RT)-PCR kit (Takara) containing SYBR green dye. Each reaction was performed in triplicate. Relative expression level of hACE2 and mACE2 was determined using primer pairs ehACE2 F1, ehACE2 R1 and emACE2-F1, emACE2-R1 (Table S2). Amplicons were normalized to expression of GAPDH (glyceraldehyde 3-phosphate dehydrogenase).
Western Blotting
Mouse tissues were homogenized in Cell lysis buffer for Western and IP (20 mM Tris-HCl, pH 7.5, containing 150 mM NaCl, 100 mM EDTA, and 1% Triton X-100) supplemented with Phenylmethanesulfonyl fluoride (PMSF) (Beyotime). The denatured protein lysates were separated using 10% SDS-PAGE gels. After transfer, ACE2 antibody (Huabio, 1:1000) and GAPDH antibody (Abcam, 1:1000) were added, followed by horseradish peroxidase (HRP)-conjugated anti-mouse and rabbit IgG (Abcam, 1:2,000). The Pierce ECL Western Blotting Substrate (Thermo) was used for development. Bioluminescence imaging (BLI) Mice were anesthetized by intraperitoneal injection of pelltobarbitalum natricum (Sigma, 240 mg/kg body weight). tdTomato bioluminescence imaging was performed with IVIS-Lumina II imaging system(Xenogen). The relative intensities were presented as pseudocolored images as photon flux in photon (p/s/cm2/sr).
Mouse Challenge Experiments
For intranasal infection, wild-type C57BL/6 mice, young (4.5 weeks old) and aged (30 weeks old) hACE2 mice were anesthetized with sodium pentobarbital at a dose of 50 mg/kg though intraperitoneal route, and then intranasally infected with 4 × 105 pfu of SARS-CoV-2, respectively. For intragastric infection, all animals were fed with 4 × 106 pfu of SARS-CoV-2 (in 200 μl) via intragastric gavage using a ball-tipped 6-gauge feeding needle. Mice were then weighed and monitored daily, and sacrificed on day 5 or 6 post infection for serum collection and tissue processing.
Measurement of Viral RNA Loads
Viral RNA in each sample was extracted with an RNeasy Mini kit (QIAGEN) according to the protocols. The viral RNA quantification was performed by RT-qPCR targeting the S gene of SARS-CoV-2. RT-qPCR was performed using One Step PrimeScript RT-PCR Kit (Takara) with the following primers and probes: CoV-F3, CoV-R3 and CoV-P3 (Table S2).
Histopathological Analysis
Mouse tissues were excised and fixed with 10% neutral buffered formaline, dehydrated and embedded in paraffin. Each embedded tissue was sectioned into 4 μm thickness longitudinal sections. Three tissue sections derived from different parts of each tissue were stained with hematoxylin and eosin (H&E) according to standard procedures for examination by light microscopy. The degree of lung damage under the light microscopy was assessed by the degeneration of alveolar epithelial cells, the expansion of parenchymal wall, edema, hemorrhage, and inflammatory cells infiltration. The semiquantitative assessment were performed as reported previously and described briefly as the following (Haagmans et al., 2004, van den Brand et al., 2010). For the degree of degeneration of alveolar epithelial cells, we scored 0 when no alveolar epithelial cells degeneration was observed, scored 1 when the alveolar epithelial cells degeneration was less than 10%, and scored 2 when the degeneration was 10%–50%. For the degree parenchymal wall expansion, we scored 0 when no parenchymal wall expanded, scored 1 when the area of the expanded parenchymal wall was less than 10% but no focal fusion was observed, and scored 2 when the area was 10%–50% but no focal fusion was observed. For the degree of edema, we scored 0 when no edema was observed, scored 1 when the area of edema was less than 10%, and scored 2 when the area was 10%–50%. For the degree of inflammatory cells infiltration, we scored 0 when no inflammatory cell infiltration, scored 1 when occasional infiltration of single inflammatory cell was visible, and scored 2 when focal infiltration of inflammatory cells was visible. The lesion of lung was observed by an experienced experimental pathologist who focuses on the pathogenesis study of virus infection. The cumulative scores of the severity of the three sections provided the total score per animal, and the average of the three animals was taken as the total score for that group.
Multiplex Immunofluorescent Assay
3 μM paraffin sections were deparaffinised in xylene and rehydrated in a series of graded alcohols. Antigen retrievals were performed in citrate buffer (pH6) with a microwave for 20 min at 95°C followed by a 20 min cool down at room temperature. Multiplex fluorescence labeling was performed using TSA-dendron-fluorophores with NEON 7-color Allround Discovery Kit for FFPE (Histova Biotechnology). Briefly, endogenous peroxidase was quenched in 3% H2O2 for 20 min, followed by blocking reagent for 30 min at room temperature. Primary antibody was incubated for 2 h in a humidified chamber at 37°C, followed by detection using the HRP-conjugated secondary antibody and TSA-dendronfluorophores. Afterward, the primary and secondary antibodies were thoroughly eliminated by heating the slides in retrieval/elution buffer (Abcracker) for 10 s at 95°C using microwave. In a serial fashion, each antigen was labeled by distinct fluorophores. Multiplex antibody panels applied in this study are: ACE2 (Abcam, 1:200), SARS-CoV Spike (Sinobiological, 1:2000); β-IV-tubulin (Abcam, 1:1000), CC10 (Millipore, 1:500), Podoplanin (Sinobiological, 1:1000), SPC (Abcam, 1:500), CD68 (Abcam, 1:1000), Cleaved Caspase-3 (C-Casp3,CST, 1:200). After all the antibodies were detected sequentially, the slices were imaged using the confocal laser scanning microscopy platform Zeiss LSM880.
Immunohistochemistry
Paraffin-embedded tissues were sectioned at a thickness of 4 μm for immunohistochemistry staining. Sections were deparaffinized and rehydrated, endogenous peroxidases were inactivated with methanol containing 0.3% hydrogen peroxide for 30 min. Antigen retrieval was performed with citrate buffer (pH6) at 95°C for 30 min. After incubation in blocking solution (5% normal goat serum) for 10 min at room temperature, the slides were incubated with Neutrophil Marker antibody (Santa Cruz Biotechnology, 1:200) and rabbit anti- CD68 polyclonal antibody (Abcam, 1:800) overnight at 4°C. After three washes, the sections were incubated with biotinylated anti-IgG at 37°C for 1 h, followed by Streptavidin-peroxidase conjugate (Zhongshan Biotechnology). Immunoreactivity was detected using 3, 3′ diaminobenzidine and the sections were counterstained with hematoxylin for observation by microscopy. To measure the macrophage and neutrophil infiltration semiquantitatively, 10 arbitrarily chosen 40 × (neutrophil) or 100 × (macrophage) objective fields of the lung parenchyma in each lung section were examined by light microscopy for the presence of neutrophils or macrophages. This assessment was performed in a blinded fashion. The cumulative scores for each animal were expressed as the number of positive fields per 100 fields (%).
Cytokine Assay
A total of 25 μL serum from each mouse was adopt for cytokine analysis with Bio-Plex Pro Mouse Cytokine Grp I Panel 23-Plex (BIO-RAD) according to the manufacturer’s instructions. The data were collected on Luminex 200 and analyzed by Luminex PONENT (Thermo Fisher).
Quantification and Statistical Analysis
Statistical analyses were carried out using Prism software (GraphPad Prism 7.0). Data of semiquantitative analysis of the H&E-stained lung sections are presented as mean, and the statistical significance between the WT and hACE2 mice was assessed by Mann Whitney test of Student’s t tests. The other data are presented as mean ± sem, and the statistical significance of IHC staining analysis between the WT and hACE2 mice was assessed by unpaired Student’s t tests. Before the statistical significance was assessed, Shapiro-Wilk normality test was used to determine whether the data met normal distribution. Statistical details of experiments and animal replication numbers (n) are stated in the relevant figure legends and method details. A p value less than 0.05 is considered statistically significant.
Acknowledgments
This work was supported by the National Key Research and Development Project of China, China (2016YFD0500304, 2020YFA0707801); the National Science and Technology Major Project of China, China (No. 2017ZX103304402, 2018ZX09711003); the National Natural Science Foundation of China, China (82041006, 31871476); and Guangdong Pearl River talent plan, China (2019CX01N111). C.F.Q. was supported by the National Science Fund for Distinguished Young Scholar, China (No. 81925025); the Innovative Research Group (No. 81621005) from the National Natural Science Foundation of China, China; and the Innovation Fund for Medical Sciences (No. 2019-I2M-5-049) from the Chinese Academy of Medical Sciences, China.
Author Contributions
C.F.Q., C.F.F., Y.S.Z., and Y.C.W. conceived this project. S.H.S., Q.C., H.J.G., G.Y., Y.X.W., X.Y.H., S.S.L., N.N.Z., X.F.L., Y.G., and Y.Q.D. performed the experiments and analyzed all the results. W.J.H., Q. L., Q.M.L., Y.L.S., Y.Z., X.Y., and T.Y.Z. helped analyze the results. Y.C.W., C.F.Q., Y.S.Z., X.Y., and C.C.F. co-supervised the project. C.F.Q., Q.C., Y.S.Z., Y.C.W., and C.F.F. wrote and revised the manuscript with input from all authors.
Declaration of Interests
The authors declare no competing interests.
Published: May 27, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.chom.2020.05.020.
Contributor Information
Chang-Fa Fan, Email: fancf@nifdc.org.cn.
Yu-Sen Zhou, Email: yszhou@bmi.ac.cn.
Cheng-Feng Qin, Email: qincf@bmi.ac.cn.
You-Chun Wang, Email: wangyc@nifdc.org.cn.
Supplemental Information
References
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020 doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Cai J., Xu J., Lin D., Yang Z., Xu L., Qu Z., Zhang Y., Zhang H., Jia R., Liu P. A Case Series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa198. ciaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.H., Peiris J.S., Lam S.Y., Poon L.L., Yuen K.Y., Seto W.H. The Effects of Temperature and Relative Humidity on the Viability of the SARS Coronavirus. Adv. Virol. 2011;2011 doi: 10.1155/2011/734690. 734690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV--a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagmans B.L., Kuiken T., Martina B.E., Fouchier R.A., Rimmelzwaan G.F., van Amerongen G., van Riel D., de Jong T., Itamura S., Chan K.H. Pegylated interferon-alpha protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat. Med. 2004;10:290–293. doi: 10.1038/nm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Lian J.S., Hu J.H., Gao J., Zheng L., Zhang Y.M., Hao S.R., Jia H.Y., Cai H., Zhang X.L. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69:1002–1009. doi: 10.1136/gutjnl-2020-320926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Jiang X., Zhang Z., Huang S., Zhang Z., Fang Z., Gu Z., Gao L., Shi H., Mai L. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69:997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. USA. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netland J., Meyerholz D.K., Moore S., Cassell M., Perlman S. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J. Virol. 2008;82:7264–7275. doi: 10.1128/JVI.00737-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., SARS study group Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F., Qian S., Zhang S., Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020;526:135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C.T., Huang C., Newman P., Wang N., Narayanan K., Watts D.M., Makino S., Packard M.M., Zaki S.R., Chan T.S., Peters C.J. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human Angiotensin-converting enzyme 2 virus receptor. J. Virol. 2007;81:1162–1173. doi: 10.1128/JVI.01702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brand J.M., Stittelaar K.J., van Amerongen G., Rimmelzwaan G.F., Simon J., de Wit E., Munster V., Bestebroer T., Fouchier R.A., Kuiken T., Osterhaus A.D. Severity of pneumonia due to new H1N1 influenza virus in ferrets is intermediate between that due to seasonal H1N1 virus and highly pathogenic avian influenza H5N1 virus. J. Infect. Dis. 2010;201:993–999. doi: 10.1086/651132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Bushmaker T., Munster V.J. Stability of Middle East respiratory syndrome coronavirus (MERS-CoV) under different environmental conditions. Euro Surveill. 2013;18:20590. doi: 10.2807/1560-7917.es2013.18.38.20590. [DOI] [PubMed] [Google Scholar]
- van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yang B., Li Q., Wen L., Zhang R. Clinical Features of 69 Cases with Coronavirus Disease 2019 in Wuhan, China. Clin. Infect. Dis. 2020:ciaa272. doi: 10.1093/cid/ciaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.H., Deng W., Tong Z., Liu Y.X., Zhang L.F., Zhu H., Gao H., Huang L., Liu Y.L., Ma C.M. Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comp. Med. 2007;57:450–459. [PubMed] [Google Scholar]
- Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020;5:335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.