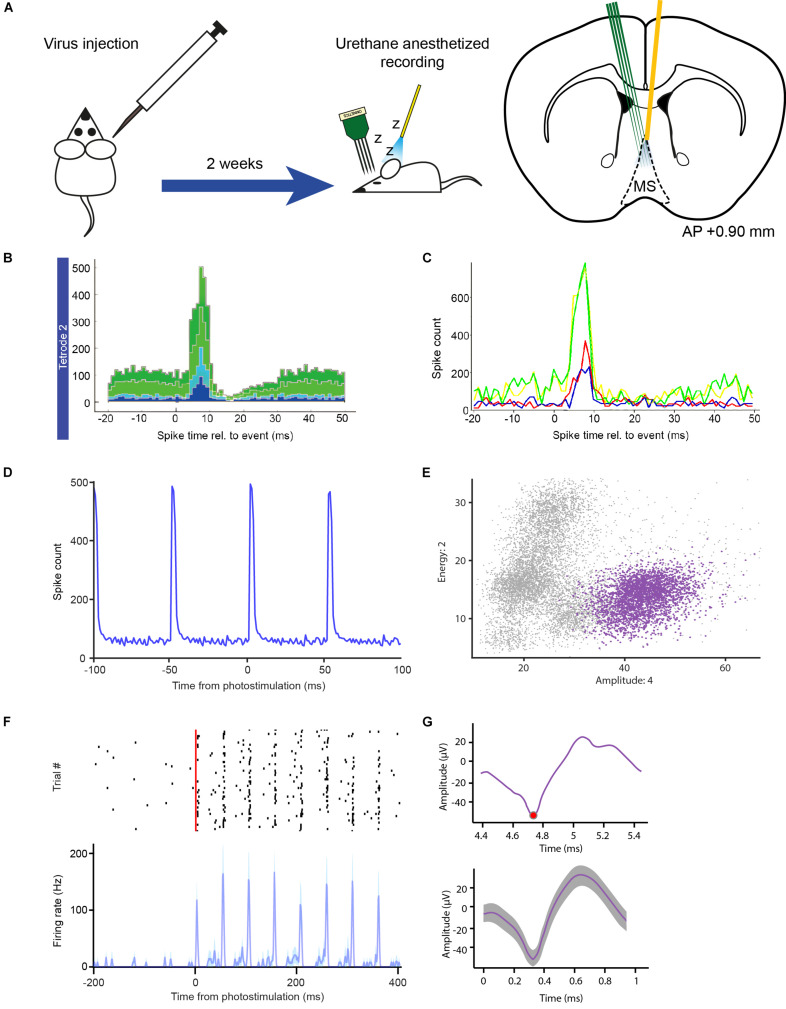
FIGURE 6.
In vivo optogenetic tagging experiment for testing OPETH. (A) Transgenic animals were virus injected and, after a 2 weeks recovery period, acute recordings were performed under urethane anesthesia, in which a 32-channels silicon probe and an optic fiber were inserted in the MS in order to record light-activated units. (B) OPETH Histogram Window (“aggregate view”) shows neural responses to laser pulses in a Vglut2-Cre mouse expressing channelrhodopsin2 (1 ms bin size). (C) OPETH “Channel view” of the same recording was used to determine the most responsive channel of the tetrode of interest. In this view, histograms are calculated separately for single channels (arbitrary color code). (D) The offline peri-event time histogram calculated from all spikes of the same tetrode (unsorted data of all neurons) confirmed the presence of strong light-evoked activity. (E) Spikes from the same tetrode were sorted into putative single neurons using MClust 3.5. The plot shows an optotagged neuron in feature space (purple). Note the relatively small deviations around the cluster centroid, characteristic to real spikes in contrast with light-evoked artifacts. (F) Spike raster (black, spike times) and peri-event time histogram of the sorted MS single neuron example aligned to the onset of the laser pulse train (red), from the channel selected via OPETH (resolution, 1 ms). (G) Top, raw light-evoked spike in the OPETH Spike window. Bottom, average spike shape of the same neuron analyzed offline.