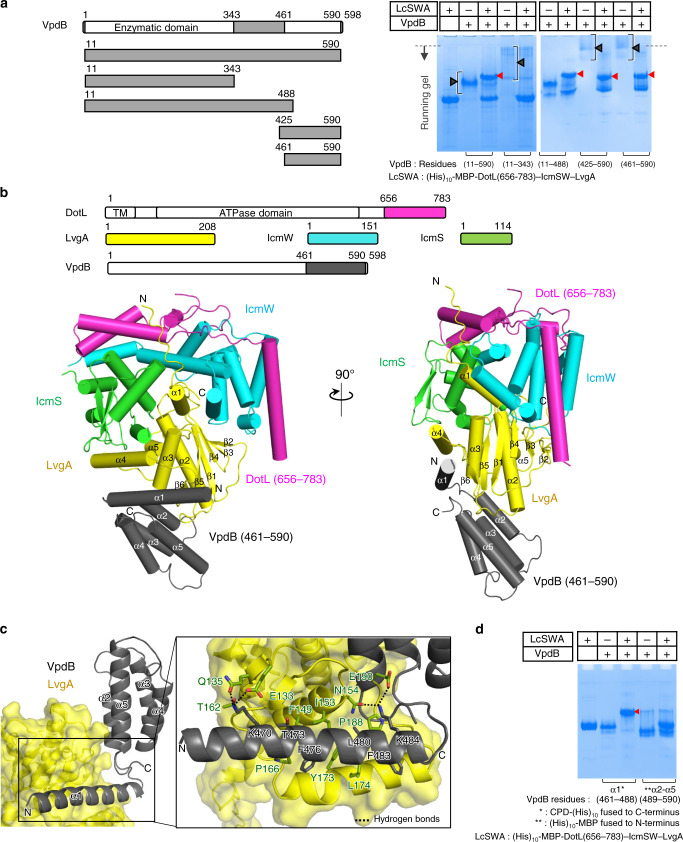
Fig. 1. Crystal structure of DotL(656-783)–IcmSW–LvgA–VpdB(461-590).
a VpdB(461-590) binds to (His)10-MBP-DotL(656-783)–IcmSW–LvgA. Each of the five truncated VpdB proteins (10 μM) was incubated with (His)10-MBP-DotL(656-783)–IcmSW–LvgA at a 1:1 molar ratio and visualized on a native PAGE-gel by Coomassie staining. Schematic drawings of the constructs of VpdB are shown on the left. The black brackets and the red arrowheads indicate the diffusive input protein bands and newly formed protein bands, respectively. The dotted line indicates the borderline between the stacking and the running gel. b Two views of structure of the DotL(656-783)–IcmSW–LvgA–VpdB(461-590). Schematic drawings of the five protein constructs are shown at the top. Protein constructs used for structure determination are color coded and labeled. c Enlarged view of the interaction between VpdB(461-590) and LvgA. α1 of VpdB(461-590) lies on a wide and shallow groove of LvgA. VpdB residues majorly involved in the intermolecular interaction are shown as sticks. d Helix α1 is the major binding motif. (His)10-MBP-DotL(656-783)–IcmSW–LvgA (10 μM) was incubated with CPD-(His)10 tagged helix α1 or (His)10-MBP tagged four-helix bundle α2–α5 of VpdB(461-590) at a 1:1 molar ratio, and visualized on a native gel. Only helix α1 exhibited a newly formed protein band indicated by the red arrowhead. All the native PAGE analyses were repeated more than 3 times.