The Phe43 cavity of HIV-1 envelope glycoproteins (Env) is an attractive druggable target. New promising compounds, including small CD4 mimetics (CD4mc), were shown to insert deeply into this cavity. Here, we identify a new network of residues that helps to shape this highly conserved CD4 binding pocket and characterize the structural determinants responsible for Env sensitivity to small CD4 mimetics.
KEYWORDS: Env, gp120, CD4, CD4mc, CRF01_AE, ADCC, CD4 binding site, CD4 mimetic, HIV-1, Phe43 cavity, neutralization
ABSTRACT
The HIV-1 envelope glycoproteins (Env) undergo conformational changes upon interaction of the gp120 exterior glycoprotein with the CD4 receptor. The gp120 inner domain topological layers facilitate the transition of Env to the CD4-bound conformation. CD4 engages gp120 by introducing its phenylalanine 43 (Phe43) in a cavity (“the Phe43 cavity”) located at the interface between the inner and outer gp120 domains. Small CD4-mimetic compounds (CD4mc) can bind within the Phe43 cavity and trigger conformational changes similar to those induced by CD4. Crystal structures of CD4mc in complex with a modified CRF01_AE gp120 core revealed the importance of these gp120 inner domain layers in stabilizing the Phe43 cavity and shaping the CD4 binding site. Our studies reveal a complex interplay between the gp120 inner domain and the Phe43 cavity and generate useful information for the development of more-potent CD4mc.
INTRODUCTION
The human immunodeficiency virus type 1 (HIV-1) envelope glycoproteins (Env) mediate virus entry into host cells to initiate the viral replication cycle. The gp120 exterior Env subunit mediates the initial interaction with the CD4 receptor. The gp120 phenylalanine 43 (Phe43) cavity is a conserved region of ∼150 Å3 where Phe43 of CD4 makes numerous contacts with conserved gp120 residues and is connected with the gp120 inner domain and the coreceptor binding site via a water-filled solvent channel (1, 2). This interaction triggers major conformational changes allowing coreceptor (i.e., CCR5 and CXCR4) binding (3–10). Subsequent conformational changes in gp41 lead to the formation of the six-helix bundle, resulting in the fusion of the viral envelope and the target cell membrane (11, 12). CD4-induced (CD4i) conformational changes in the gp120 inner domain involve three flexible topological layers (layers 1, 2, and 3). Despite a lack of contact with CD4, the gp120 inner domain layers govern CD4 triggering by participating in conformational transitions within gp120 and regulating the interaction with gp41 (13, 14). Structural rearrangements between layer 1 and layer 2 were previously shown to facilitate the transition of the envelope glycoprotein trimer from the unliganded state to the CD4-bound state and to stabilize gp120-CD4 interactions (14–16). Layer 3 was previously shown to govern the efficiency of the initial gp120 interaction with CD4, a function that can also be fulfilled by filling the Phe43 cavity with a bulky residue at position 375 (13, 14, 17).
Env represents the only viral antigen exposed on the surface of virions and infected cells and thus is the primary target for antibodies (Abs). Despite targeting different epitopes, most broadly neutralizing antibodies (bNAbs) preferentially recognize the “closed” conformation of Env (18, 19). Conversely, CD4 binding forces Env to adopt “open” conformations (20, 21), allowing its recognition by nonneutralizing Abs (nnAbs) (22, 23). Env-CD4 complexes at the surface of HIV-1-infected cells were previously shown to be efficiently recognized by CD4-induced (CD4i) Abs, which are present in sera from HIV-1-infected individuals and can mediate potent antibody-dependent cellular cytotoxicity (ADCC) responses (24–28). HIV-1 evolved to minimize the exposure of these CD4i epitopes by limiting Env-CD4 interaction. HIV-1 accomplishes this through its Nef and Vpu accessory proteins, which decrease the overall amounts of Env (via Vpu-mediated BST-2 downregulation) and CD4 at the cell surface (23, 29, 30). In addition, efficient Env internalization also limits ADCC responses (31, 32). Motivated by the vulnerability of the CD4-bound conformation of Env to ADCC, new approaches to “force” Env to adopt this conformation using small CD4-mimetic compounds (CD4mc) have been developed (33). The antiviral activity of CD4mc is not limited to ADCC; these compounds can compete for Env-CD4 interaction, mediate viral particle inactivation by prematurely triggering Env, and sensitize infectious viral particles to neutralization by otherwise nnAbs (34–37). The potential clinical benefit of CD4mc was highlighted in two recent in vivo studies showing how CD4mc can act as prophylactic agents to decrease HIV-1 acquisition in humanized mice and simian-human immunodeficiency virus (SHIV)-challenged nonhuman primates (NHP) (38, 39).
Interestingly, Env transitions to the CD4-bound conformation can be modulated by single-residue substitutions. For example, the replacement of the well-conserved group M serine at position 375 by a large hydrophobic residue, such as tryptophan, fills the Phe43 cavity; this substitution alters Env conformation by predisposing gp120 to spontaneously assume a state closer to the CD4-bound conformation (13, 14, 40). While S375 is well conserved in the majority of group M HIV-1 isolates, CRF01_AE Env possesses a Phe43 cavity-filling residue at position 375 (H375) (41–43). The presence of H375 was linked to the natural exposure of CD4i epitopes in CRF01_AE strains, resulting in their enhanced susceptibility to ADCC responses (42). Besides modulating Env interaction with human CD4 (41), residue 375 was shown to modulate SHIV binding to rhesus monkey CD4 and replication in nonhuman primates (44), highlighting its critical role in viral pathogenesis. By performing structural, in silico, and functional analyses, using CD4mc as probes, we uncovered how the gp120 inner domain shapes the Phe43 cavity and the CD4 binding site.
RESULTS
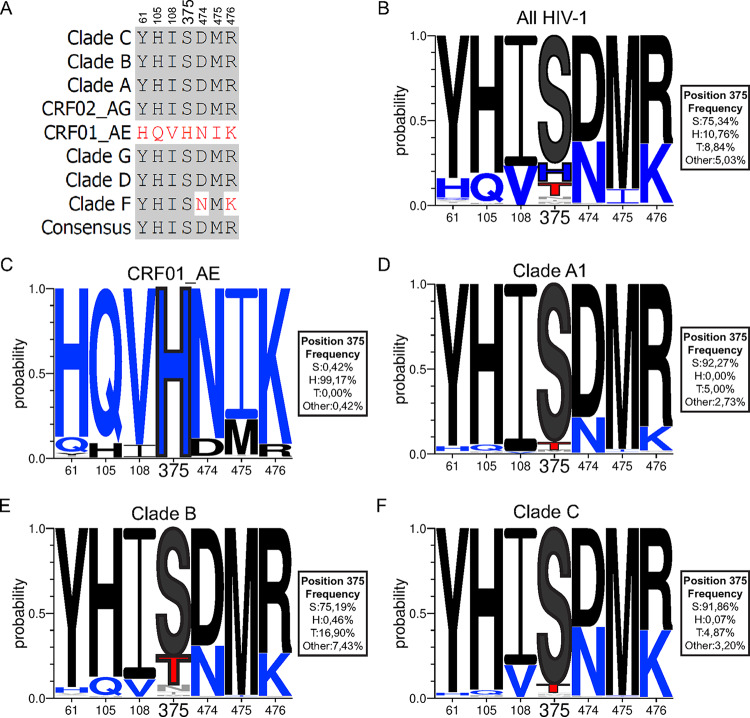
Comparison of Phe43 cavity and coevolving inner domain layer residues among HIV-1 clades.
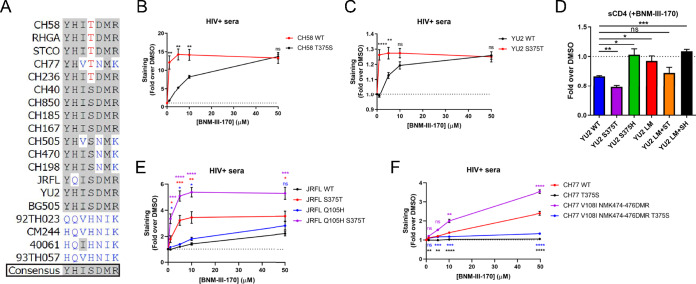
We previously reported that six residues within the gp120 inner domain layers coevolved with Phe43 cavity residue 375 to facilitate CD4 interaction (41). We analyzed all available HIV-1 sequences together or segregated by clades using the NIH Los Alamos HIV database to determine the degree of conservation of these residues located in layer 1 (residue 61), layer 2 (residues 105 and 108), and layer 3 (residues 474, 475, and 476) (collectively named LM, for layer mutants) and in the Phe43 cavity (residue 375). The consensus sequences of these residues from CRF01_AE strains diverge from those of all other major HIV-1 subtypes (clades A, B, C, D, F, G, and CRF02_AG) (Fig. 1A). Among the coevolving inner domain residues, most of the clades (except clade F [N474 and K476]) share the same consensus sequence (Y61, H105, I108, D474, M475, and R476), which differs from that seen with the residues found in CRF01_AE strains (H/Q61, Q105, V108, N474, I475, K476). Serine 375 is the predominant residue (>75%) in all HIV-1 major subtypes, except for CRF01_AE (Fig. 1A to C). Other residues were found to occupy position 375 (T375, N375, I375, and M375), with T375 being present in more than 8% of all HIV-1 strains (Fig. 1B). T375 is present in clade B (16.9%), clade A1 (5%), and clade C (4.87%) but also in clade D (7.76%), CRF02_AG (5.26%), and clade F (2.27%). Interestingly, CRF01_AE strains have a highly conserved histidine at position 375 (H375; >99%) (41–43).
FIG 1.
Sequence alignment of gp120 Phe43 cavity and inner domain layer residues of different HIV-1 isolates. (A) Sequence alignment of selected gp120 residues located in the Phe43 cavity (375) or the inner domain layers (61, 105, 108, 474, 475, and 476) based on the Env consensus sequence of each of the HIV-1 group M major subtypes (clades A, B, C, D, F, G, CRF01_AE, and CRF02_AG). The 2017 Los Alamos database-curated Env alignment was used as the basis for this figure, which contains 5,471 amino acid HIV-1 group M sequences (including 481 sequences of CRF01_AE, 220 of subtype A1, 1,937 of subtype B, and 1,377 of subtype C). Residue numbering is based on that of the HXBc2 strain of HIV-1. Identical residues are shaded in dark gray, and nonidentical residues are highlighted in red. (B to F) Logo depictions of the frequency of each amino acid from the Phe43 cavity at positions 61, 105, 108, 375, 474, 475, and 476 in all HIV-1 isolates (B) or in isolates from CRF01_AE (C), clade A1 (D), clade B (E), and clade C (F). The height of the letter indicates its frequency within the clade. S375 and its coevolving residues are shown in black, H375 and its coevolving residues are shown in blue, and T375 is shown in red. The box beside each logo indicates the frequency of all the amino acids at position 375.
Phe43 cavity and gp120 inner domain layer changes render CRF01_AE strains susceptible to CD4mc antiviral activities.
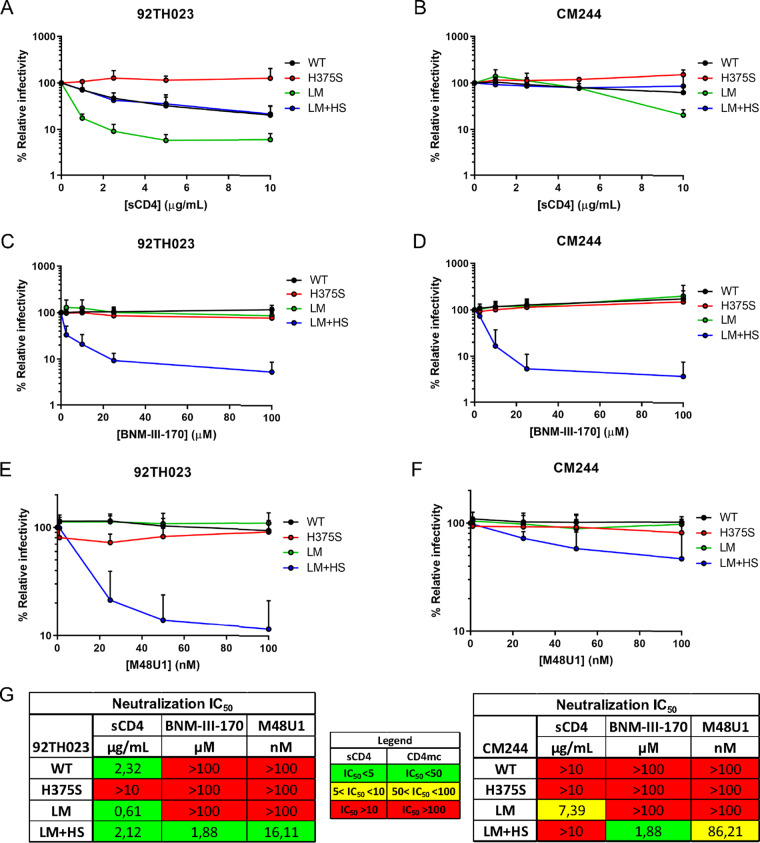
To evaluate the potential impact of the LM residues on shaping the Phe43 cavity, we decided to use CD4mc as probes. Several cocrystals of different CD4 mimics and gp120 cores exist, providing detailed structural information regarding their mode of interaction within the cavity (1, 37, 45–51). Briefly, both CD4mc (molecular weight [MW], ∼500 Da) and CD4 miniproteins (∼3 kDa) were optimized to display a large hydrophobic group (phenyl or cyclohexyl ring) that projects deeply into the Phe43 cavity, allowing them to reach the cavity edge where residue 375 is located. These projections were found to go deeper into the Phe43 cavity than CD4 Phe43 residue itself, rendering these molecules more sensitive to changes occurring within the cavity. It has been previously reported that the presence of bulky cavity-filling residues at this position (H375, W375, Y375, R375) confers resistance to CD4mc and CD4 miniproteins by abrogating their interaction deep within the Phe43 cavity (34, 36, 47, 48, 50, 52, 53). Replacement of the naturally occurring CRF01_AE H375 with a serine residue (H375S) has been shown to reduce CD4 binding. Introduction of the LM substitutions (H/Q61Y, Q105H, V108I, N474D, I475M, and K476R) restored this interaction (41). However, whether these Phe43 cavity and inner domain changes affect CD4mc sensitivity has not yet been determined. First, we evaluated the effect of the H375S change on the sensitivity of two CRF01_AE isolates (tier 1 92TH023 and tier 2 CM244) to neutralization by different CD4 mimics, including soluble CD4 (sCD4), CD4mc (BNM-III-170), and a CD4 miniprotein (M48U1) (33, 54). In agreement with previous results (41), replacement of histidine 375 by a serine residue (H375S) in both HIV-1CRF01_AE Envs completely abolished the susceptibility of pseudotyped virions to sCD4 neutralization (Fig. 2A and B to G). Viral particles bearing both CRF01_AE Envs were resistant to neutralization by BNM-III-170 and M48U1; this was expected due to the presence of the cavity-filling histidine at position 375 (H375). Interestingly, replacement of the bulky histidine by a serine at this position (H375S) did not restore neutralization sensitivity to these CD4 mimics (Fig. 2C to G). However, the H375S change in combination with the LM mutations (LM plus HS [LM+HS]) dramatically enhanced the susceptibility of both CRF01_AE strains to neutralization by both CD4 mimics. Of note, the presence of different combinations of single-layer or multiple-layer changes together with the H375S change was not sufficient to restore sensitivity to neutralization by BNM-III-170 or M48U1 (see Fig. S1C to G in the supplemental material). This was different from the results seen with respect to sCD4 neutralization, where viruses bearing LM changes without the Q61Y change were also sensitive to sCD4 neutralization (Fig. S1A and G). These data highlight subtle differences in the mode of CD4mc and sCD4 recognition of Env. Altogether, these results indicate that all of the LM changes are required to restore CD4mc sensitivity to CRF01_AE strains presenting an “empty” (H375S) Phe43 cavity.
FIG 2.
Effect of gp120 layer mutations on neutralization by CD4 and CD4mc. Recombinant HIV-1 strains expressing luciferase and bearing wild-type or mutant CRF01_AE Envs (92TH023 and CM244 isolates) were normalized by reverse transcriptase activity. Normalized amounts of viruses were incubated with serial dilutions of sCD4 (A and B), BNM-III-170 (C and D), or M48U1 (E and F) at 37°C for 1 h prior to infection of Cf2Th-CD4/CCR5 cells. Infectivity at each dilution of sCD4 or CD4mc tested is shown as the percentage of infection without sCD4 or CD4mc for each particular mutant. Quadruplicate samples were analyzed in each experiment. Data shown are the means of results obtained in at least 3 independent experiments. The error bars represent the standard deviations. Neutralization half-maximal inhibitory concentration (IC50) values are summarized in panel G.
Effect of single gp120 layer mutations on neutralization by CD4 and CD4mc. Recombinant HIV-1 strains expressing luciferase and bearing wild-type or mutant CRF01_AE Envs (92TH023 and CM244 isolates) were normalized by reverse transcriptase activity. Normalized amounts of viruses were incubated with serial dilutions of sCD4 (A and B), BNM-III-170 (C and D), or M48U1 (E and F) at 37°C for 1 h prior to infection of Cf2Th-CD4/CCR5 cells. Infectivity at each dilution of sCD4 or CD4mc tested is shown as the percentage of infection without sCD4 or CD4mc for each particular mutant. Quadruplicate samples were analyzed in each experiment. Data shown are the means of results obtained in at least 3 independent experiments. The error bars represent the standard deviations. Neutralization half-maximal inhibitory concentrations (IC50) are summarized in panel G. Download FIG S1, TIF file, 0.5 MB (481.8KB, tif) .
Copyright © 2020 Prévost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
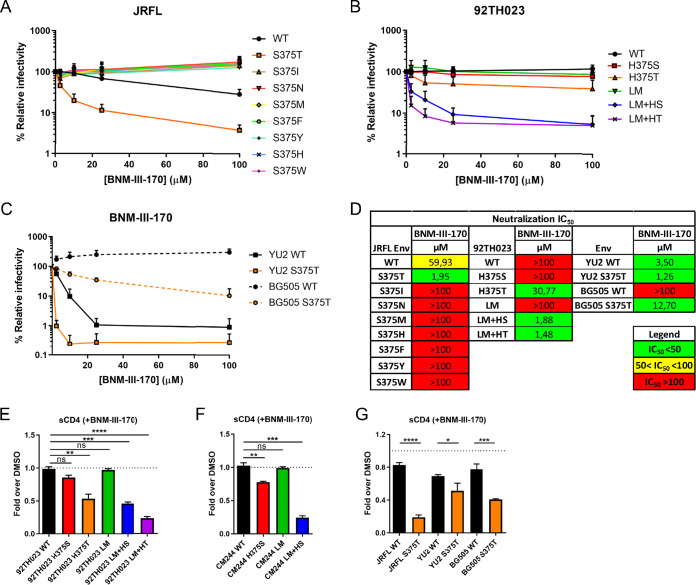
As a small residue at position 375 such as serine appears to be required for CD4mc sensitivity, we investigated the possibility that another naturally occurring small residue such as threonine (T), isoleucine (I), or asparagine (N) might affect HIV-1 sensitivity to CD4mc. We first introduced these changes into clade B HIV-1JRFL Env and performed neutralization assays using the CD4mc BNM-III-170. Among all the residue 375 substitutions tested, T375 was found to be more sensitive to BNM-III-170 inhibition than the wild type (WT) (S375) and all the other variants (Fig. 3A). We then introduced this change into a CRF01_AE Env alone (H375T) and in combination with the six layer mutations (LM+HT). Strikingly, the H375T change was sufficient to sensitize the CRF01_AE strain to BNM-III-170 neutralization in the absence of the LM mutations. Thus, despite having similar small side chains, T375 was found to be sufficient to sensitize the CRF01_AE virus to neutralization by BNM-III-170 whereas S375 required the LM mutations to do so. Addition of the LM changes together with H375T (LM+HT) further increased the sensitivity of the CRF01_AE virus to BNM-III-170 neutralization, resulting in levels comparable to those seen with the LM+HS mutant (Fig. 3B and D). We confirmed the enhanced susceptibility of T375-bearing viruses to BNM-III-170 with additional clade B (YU2) and clade A1 (BG505) strains. Increased neutralization by CD4mc was seen in the presence of T375 in these two strains (Fig. 3C and D). Strikingly, HIV-1BG505 has intrinsic resistance to CD4mc BNM-III-170 which can be overcome by the presence of T375.
FIG 3.
Effect of Env residue 375 on CD4mc neutralization sensitivity. (A to D) Recombinant HIV-1 strains expressing luciferase and bearing wild-type or mutant Envs (JRFL, 92TH023, YU2, and BG505 isolates) were normalized by reverse transcriptase activity. Normalized amounts of viruses were incubated with serial dilutions of BNM-III-170 (A to C) at 37°C for 1 h prior to infection of Cf2Th-CD4/CCR5 cells. Infectivity at each dilution of CD4mc tested is shown as the percentage of infection without CD4mc for each particular mutant. Quadruplicate samples were analyzed in each experiment. Data shown are the means of results obtained in at least 3 independent experiments. The error bars represent the standard deviations. Neutralization half-maximal inhibitory concentrations (IC50) are summarized in panel D. (E to G) Cell surface staining of 293T cells transfected with different Env expressors (92TH023, CM244, JRFL, YU2, and BG505 isolates [WT or their mutated counterparts]). Binding of sCD4 in the presence of BNM-III-170 (50 μM) or in its absence (DMSO) was detected with the anti-CD4 OKT4 MAb. Shown are the mean fluorescence intensities (MFI) obtained in the presence of BNM-III-170 normalized to the MFI in the absence of BNM-III-170 (DMSO) from the transfected (GFP+) population for staining obtained in at least 3 independent experiments. All MFI data were normalized to 2G12 MFI for each Env mutant. Error bars indicate means ± standard errors of the means (SEM). Statistical significance was tested using an unpaired t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant).
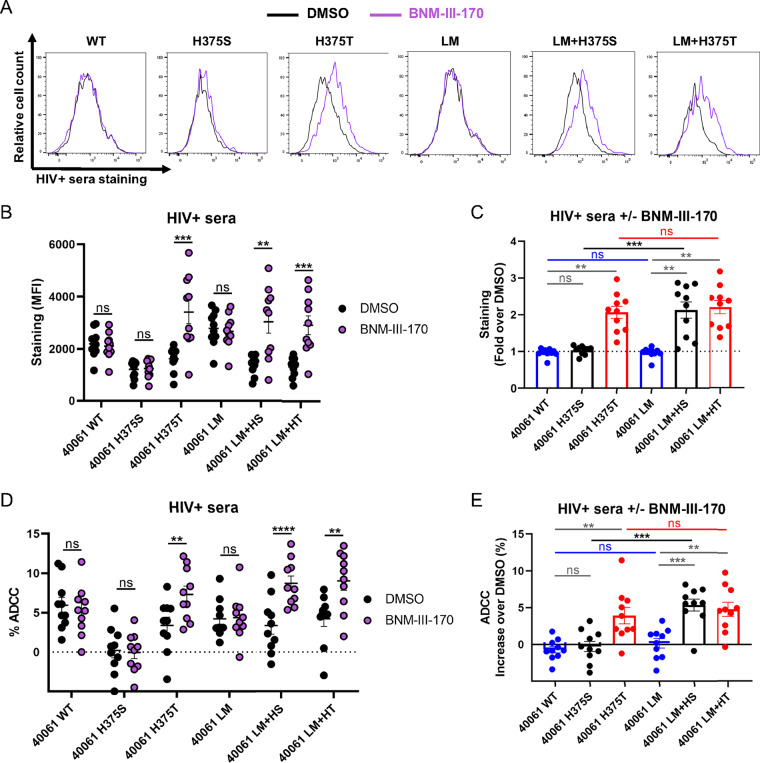
Since CD4mc were also shown to sensitize HIV-1-infected cells to ADCC by CD4-induced (CD4i) Abs present in HIV-positive (HIV+) sera (27, 33, 55–57), we evaluated the impact of the Phe43 and LM changes on the susceptibility of CRF01_AE-infected primary CD4+ T cells to ADCC mediated by HIV+ sera in the presence of the CD4mc BNM-III-170. Briefly, primary CD4+ T cells cultured from uninfected individuals were infected with the wild-type (WT) CRF01_AE transmitted/founder (TF) 40061 strain or with an isogenic virus with the LM and 375 changes. This infectious molecular clone (IMC) was isolated from a participant in the acute HIV-1 infection RV217 cohort (58). In line with the results presented in Fig. 2 and 3 (see also Fig. S1), replacement of CRF01_AE H375 with a threonine, but not a serine, was sufficient to gain sensitivity to CD4mc, leading to an increase in binding and ADCC mediated by HIV+ sera (Fig. 4). Addition of the LM changes to the resistant H375S variant conferred BNM-III-170 sensitivity but did not further enhance the susceptibility of the H375T variant.
FIG 4.
Phe43 cavity and inner domain changes enhance susceptibility of the CRF01_AE HIV-1 strain to ADCC responses mediated by HIV+ sera in the presence of CD4mc. (A to C) Cell surface staining of primary CD4+ T cells infected with a CRF01_AE transmitted-founder (TF) strain (40061) and variants with 10 different HIV+ sera. (A) Histograms depicting representative HIV+ serum staining. (B and C) The graphs shown represent the compiled mean fluorescence intensities (MFI) obtained with 10 HIV+ sera (B) and the fold increase in MFI in the presence of CD4mc calculated for the infected (p24+) population (C). (D and E) Infected primary CD4+ T cells were also used as target cells, and autologous PBMCs were used as effector cells in a FACS-based ADCC assay. The graphs shown represent (D) the ADCC values and (E) the increase (in percentage points) of ADCC levels obtained in the presence of CD4mc with 10 HIV+ sera. These results were obtained in at least 2 independent experiments. Error bars indicate means ± SEM. Statistical significance was tested using a paired t test or a Wilcoxon rank test based on statistical normality (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant).
To gain a better understanding of the impact that the LM and 375 changes have on Env conformation, we developed an assay to indirectly measure the capacity of BNM-III-170 to interact with Env. Briefly, CD4-negative cells were transfected with HIV-1CRF01_AE 92TH023 or CM244 Env variants. At 48 h posttransfection, Env-expressing cells were incubated with sCD4 in the presence of the CD4mc BNM-III-170 or an equivalent amount of the vehicle (dimethyl sulfoxide [DMSO]). Binding of sCD4 to cell surface Env was detected using an anti-CD4 MAb (OKT4), which recognizes the CD4 D3 domain and therefore does not compete for gp120 binding. The ability of BNM-III-170 to interact with Env was inferred by calculating the decrease in the level of sCD4 binding to Env in the absence of CD4mc (DMSO) or in its presence (Fig. 3E to G). In agreement with neutralization results, the occupancy of the Phe43 cavity of CRF01_AE strains by BNM-III-170 was increased as a consequence of a combination of Phe43 cavity and inner domain changes (LM+HS and LM+HT) or of the H375T mutation change alone. We observed a decrease of at least 50% in sCD4 binding for both CRF01_AE strains with these mutants (Fig. 3E and F). Similarly, CD4mc competed more efficiently for sCD4 binding in the presence of the S375T change for the three additional Envs tested (JRFL, YU2, BG505) (Fig. 3G). Using this assay, we also tested a panel of anti-Env antibodies, where bNAbs (3BNC117, NIH45-46 G54W, PG16, PGT151, and PGT128) preferentially recognize the “closed” trimer (18, 19, 59) and nnAbs (17b, 19b, F240, and A32) preferentially recognize the “open” trimer (18, 19, 27, 60). In agreement with the higher level of occupancy of the Phe43 cavity by BNM-III-170 in the presence of T375 or the LM+HS mutations, for these mutants, treatment of Env-expressing cells with the CD4mc BNM-III-170 more effectively decreased binding of bNAbs to the “closed” Env conformation (Fig. S2A and B and E to G) and concomitantly enhanced binding of nnAbs to the “open” conformation (Fig. S2C and D and H to J). Overall, the nature of the residue 375 and its coevolving inner domain residues regulate the susceptibility of the CRF01_AE strains to both neutralization and ADCC.
Phe43 cavity and inner domain changes render the CRF01_AE HIV-1 strain susceptible to CD4mc-induced Env conformational changes. Cell surface staining of 293T cells transfected with different Env expressors (92TH023, CM244, JRFL, YU2, and BG505 isolates [WT or their mutated counterparts]) was performed using a panel of Env ligands. Binding of bNAbs (panels A and B and panels E to G) and nnAbs (panels C and D and panels H to J) was performed in the presence of BNM-III-170 (50 μM) or in its absence (DMSO). Shown are the mean fluorescence intensities (MFI) obtained in the presence of BNM-III-170 normalized to the MFI or in absence of BNM-III-170 (DMSO) from the transfected (GFP+) population for staining obtained in at least 3 independent experiments. All MFI data were normalized to 2G12 MFI for each Env mutant. Error bars indicate means ± SEM. Statistical significance was tested using an unpaired t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant). Download FIG S2, TIF file, 0.5 MB (473.4KB, tif) .
Copyright © 2020 Prévost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
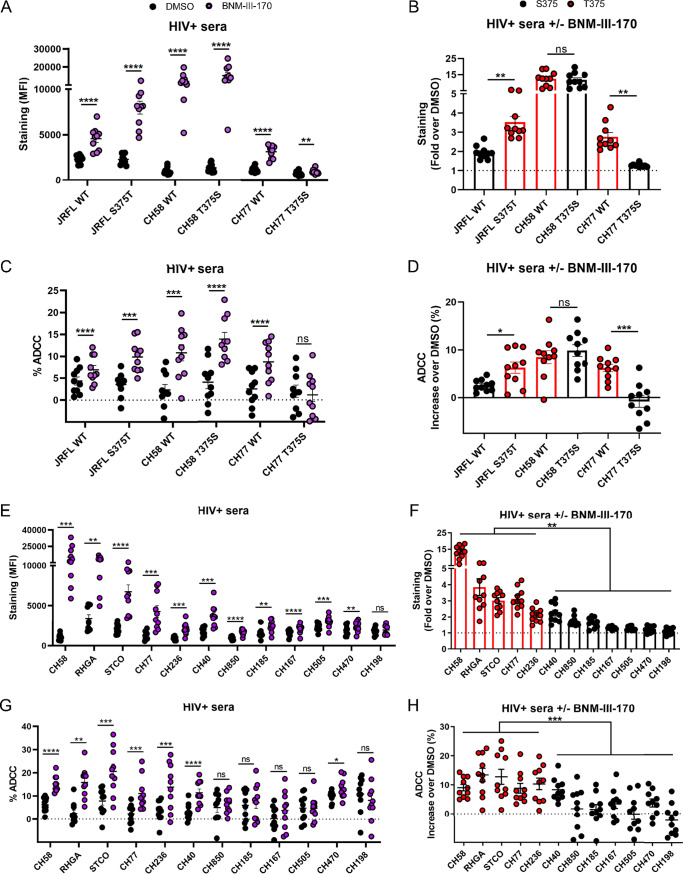
Phe43 cavity and layer residues modulate the sensitivity of HIV-1 of major clades to CD4mc antiviral activities.
In contrast with CRF01_AE strains, most strains from pandemic HIV-1 major clades (clades A, B, C, D, G, and CRF02_AG) harbor inner domain residues that coevolved with the presence of a small residue at position 375 (e.g., S375), making them potentially sensitive targets for CD4mc antagonist action. The presence of naturally occurring T375 in a significant fraction of these clades (up to 16.9% in clade B) might affect the susceptibility of these strains to CD4mc. Considering that T375 was found to be the residue that conferred the highest sensitivity to CD4mc in the clade B JRFL strain (Fig. 3) and among all CRF01_AE strains tested (Fig. 3 and 4), we introduced the T375 change into the HIV-1JRFL IMC and evaluated the capacity of BNM-III-170 to expose epitopes recognized by sera from 10 HIV-1-infected individuals at the surface of infected primary CD4+ T cells. In line with the neutralization data (Fig. 3), replacement of S375 with a threonine residue (S375T) significantly enhanced the recognition of JRFL-infected cells by HIV+ sera in the presence of BNM-III-170 (Fig. 5A and B). We also tested two additional primary clade B viruses (HIV-1CH58 and HIV-1CH77) that naturally harbor T375 (Fig. 6A). Replacement of T375 in these two different TF viruses with a serine (T375S) strongly reduced the sensitivity of HIV-1CH77 to CD4mc but appeared to have no effect on HIV-1CH58 (Fig. 5B). However, the CH58 T375S mutant was less sensitive to CD4mc BNM-III-170 at concentrations lower than the concentration used in the staining and ADCC assays (50 μM) (Fig. 6B). A similar phenotype was also observed with another clade B strain (HIV-1YU2), where the variant with T375 was more sensitive to BNM-III-170 at low concentrations (Fig. 6C). In all cases, a threonine residue at position 375 enhanced Env recognition by HIV+ sera in the presence of BNM-III-170 (Fig. 5A and B and 6B and C). Importantly, enhanced recognition translated into enhanced susceptibility to ADCC responses (Fig. 5C and D).
FIG 5.
Primary viruses harboring a T375 residue are highly susceptible to ADCC responses mediated by HIV+ sera in the presence of CD4mc. Primary CD4+ T cells were infected with clade B primary HIV-1 and their variants (A to D) or with a panel of TF and chronic viruses from clades B and C (E to H). At 48 h postinfection, cell surface staining was performed with 10 different HIV+ sera. The graphs shown represent (A and E) the compiled mean fluorescence intensities (MFI) obtained with 10 HIV+ sera and (B and F) the fold increase in MFI in the presence of CD4mc calculated on the infected (p24+) population. Infected primary CD4+ T cells were also used as target cells, and autologous PBMCs were used as effector cells in a FACS-based ADCC assay. The graphs shown represent (C and G) the ADCC values and (D and H) the increases (in percentage points) in the levels of ADCC obtained in the presence of CD4mc with 10 HIV+ sera. These results were obtained in at least 2 independent experiments. Error bars indicate means ± SEM. Statistical significance was tested using a paired t test or a Wilcoxon signed-rank test (A to E and G) or an unpaired t test or Mann-Whitney U test (F and H), based on statistical normality (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant).
FIG 6.
Phe43 cavity and inner domain residues modulate the susceptibility of clade B strains to CD4mc. (A) Sequence alignment of selected gp120 residues located in the Phe43 cavity (375) or the inner domain layers (61, 105, 108, 474, 475, and 476) for all HIV-1 strains used in this study. S375 and its coevolving residues are shown in black, H375 and its coevolving residues are shown in blue, and T375 is shown in red. Residues corresponding to the HIV-1 consensus sequence are shaded in gray. (B, C, E, and F) Cell surface staining with HIV+ sera of primary CD4+ T cells infected with clade B primary viruses (CH58, YU2, JRFL, and CH77) or their variants in the presence of increasing concentrations of BNM-III-170 (0, 1, 5, 10, and 50 μM). The graphs shown represent the compiled mean fluorescence intensities (MFI) obtained with 5 different HIV+ sera in the presence of BNM-III-170 normalized to the MFI in the absence of BNM-III-170 (DMSO) from the infected (p24+) population. These results were obtained in at least 2 independent experiments. (D) Cell surface staining of 293T cells transfected with an expressor of the HIV-1YU2 Env WT or its mutated counterparts. Binding of sCD4 in the presence of BNM-III-170 (50 μM) or in its absence (DMSO) was detected with the anti-CD4 OKT4 MAb. Shown are the mean fluorescence intensities (MFI) obtained in the presence of BNM-III-170 normalized to the MFI in the absence of BNM-III-170 (DMSO) from the transfected (GFP+) population for staining obtained in at least 3 independent experiments. All MFI data were normalized to 2G12 MFI for each Env mutant. Error bars indicate means ± SEM. Statistical significance was tested using an unpaired t test or Mann-Whitney U test based on statistical normality (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant).
Next, we evaluated a panel of 12 clade B and clade C primary viruses, including viruses from acute infections (transmitted/founder strains) (CH40, CH58, CH77, CH185, CH198, CH236, CH470, CH505, and CH850) and chronic infections (CH167, RHGA, STCO), for their susceptibility to BNM-III-170. We observed a significant increase in binding of HIV+ sera by BNM-III-170 with all viruses tested but one (CH198) (Fig. 5E); the increase was greater for viruses harboring a naturally occurring T375 (Fig. 5F). The higher responsiveness of viruses harboring a T375 (CH58, RHGA, STCO, CH77, and CH236) to BNM-III-170 led to a significant increase in ADCC responses for all HIV+ sera tested, whereas only a few (2/7) viruses harboring a S375 showed a significant increase in ADCC upon CD4mc addition (Fig. 5G and H). In brief, the presence of a naturally occurring T375 residue in clade B and clade C primary virus Envs increases susceptibility to CD4mc.
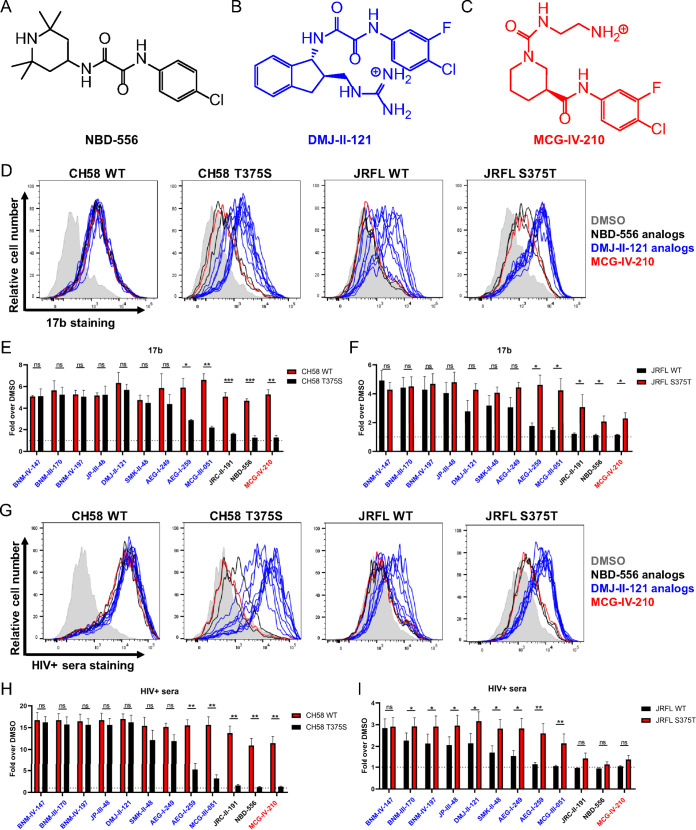
Since T375 was found to confer the highest sensitivity to BNM-III-170 in all strains tested, we sought to determine whether this was specific to this CD4mc or applicable to others as well. Therefore, we selected different generations and families of CD4mc known to engage gp120 within the Phe43 cavity. The NBD-556 compound was originally identified through a screen for Env-CD4 interaction inhibitors (61) and consists of a halogenated phenyl ring linked with a tetramethyl-piperidine via an oxalamide linker (Fig. 7A). The DMJ-II-121 compound was derived from the NBD-556 analog JRC-II-191 through the substitution of the piperidine ring for an indane ring with a guanidinium group at position 2 (Fig. 7B) to improve its Env antagonism potency (46). Other DMJ-II-121 analogs were synthesized to harbor additional chemical groups on the indane ring at position 5 (BNM-III-170, BNM-IV-147, SMK-II-48, MCG-III-051), position 6 (BNM-IV-197, JP-III-48, AEG-I-249, AEG-I-259), or position 7 (MCG-III-051) (45). To this panel of small molecules, we added a new compound (MCG-IV-210) derived from a recent high-throughput screen aimed to identify small molecules able to stabilize the CD4-bound conformation (37). Its structure resembles that of the NBD-556 analogs, but it has a shorter amide linker and an N-substituted piperidine ring (Fig. 7C). The panel of CD4mc was tested for the ability to induce Env conformational changes at the surface of primary CD4+ T cells infected with HIV-1CH58 WT or its T375S variant. Strikingly, all CD4mc tested were able to increase the exposure of the epitope recognized by the CD4i 17b MAb at the surface of WT-infected cells to similar extents (Fig. 7D to F). Introduction of the T375S change significantly diminished this ability for the less potent compounds, including MCG-IV-210, both NBD-556 analogs, and two DMJ-II-121 analogs (AEG-I-259 and MCG-III-051). The ability of these five CD4mc to enhance 17b binding to HIV-1JRFL-infected cells was increased by the replacement of S375 with a threonine residue (Fig. 7F). Similar results were obtained in experiments evaluating the ability of these CD4mc to increase the binding of HIV+ sera to CH58-infected and JRFL-infected cells (Fig. 7G to I). Altogether, our binding and functional analyses indicate that natural HIV-1 polymorphism in Ser/Thr 375 modulates Env interaction with CD4mc of different classes.
FIG 7.
Residue 375 modulates Env sensitivity to different families of CD4mc. (A to C) Chemical structures of representative members of the different families of CD4mc tested. (D to I) Cell surface staining of primary CD4+ T cells infected with CH58 or JRFL viruses and their variants with anti-Env 17b MAb (D to F) or HIV+ sera (G to I) was performed. Shown in panels D and G are histograms depicting representative 17b and HIV+ serum staining results. The graphs shown represent the fold increase in mean fluorescence intensities (MFI) in the presence of CD4mc calculated on the infected (p24+) population obtained with 17b (E and F) or with 5 different HIV+ sera (H and I). These results were obtained in at least 3 independent experiments. Error bars indicate means ± SEM. Statistical significance was tested using a paired t test or a Wilcoxon signed-rank test based on statistical normality (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonsignificant).
As shown in Fig. 2 to 4 (see also Fig. S1), specific inner domain layer residues (LM mutations) also modulate Env sensitivity to CD4mc in CRF01_AE strains. We investigated whether this could be extended to strains from other HIV-1 major clades. We replaced HIV-1 consensus inner domain residues in the clade B HIV-1YU2 Env with the residues that specifically coevolved with H375 in CRF01_AE strains (Y61H, H105Q, I108V, D474N, M475I, R476K). These changes (LM mutations) were introduced in combination with different residues at position 375 (S375, T375, or H375). Remarkably, using the sCD4 binding competition assay, the introduction of these LM changes totally abrogated the sensitivity of YU2 Env to CD4mc, but the presence of the S375T change (LM+ST) restored sensitivity to BNM-III-170 (Fig. 6D). The S375H change completely abrogated CD4mc interaction regardless of the LM mutations (Fig. 6D), as seen with CRF01_AE strains (Fig. 2, 3, and 4; see also Fig. S1). Revisiting the data shown in Fig. 5F, the three strains that were the least sensitive to BNM-III-170 were found to have inner domain residues shared with the CRF01_AE consensus sequence (Fig. 1C and 6A): CH505 (V108, N474, and K476), CH470 (N474 and K476), and CH198 (N474 and K476). Some of the LM mutations were also naturally found in two clade B strains used in this study: JRFL (Q105) and CH77 (V108, N474, and K476) (Fig. 6A). Interestingly, replacement of these residues with residues that coevolved with S375 was found to increase CD4mc sensitivity for both strains, regardless of the nature of the residue at position 375 (Fig. 6E and F). Altogether, our data show that the nature of the residue at position 375 and the coevolving gp120 inner domain layers modulate the susceptibility of HIV-1 major clades to CD4mc.
Structural analysis of the interaction of CD4mc and HIV-1 gp120 with Phe43 cavity and inner domain layer changes.
Functional analysis clearly indicates that the LM+HT Env variant is the most effective at binding CD4mc. To better understand the structural basis for the role of the LM, HS, and HT mutations in reshaping the Phe43 cavity and in CD4mc binding, we solved the crystal structures of the CRF01_AE 93TH057 gp120 extended core (gp12093TH057 coree [49]) containing the LM+HS changes in complex with two CD4mc compounds: MCG-IV-210 and BNM-III-170. The LM+HS gp12093TH057 coree–MCG-IV-210 complex was solved to 2.5-Å resolution (see Table S1 in the supplemental material; see also Fig. S3), enabling an assessment of the impact of the HS mutation by comparison to the LM+HT gp12093TH057 coree–MCG-IV-210 complex structure solved by us previously to 2.65-Å resolution (PDB identifier [ID] 6P9N) (37). We also solved the structure of the LM+HS gp12093TH057 coree–BNM-III-170 complex to 2.65-Å resolution (Table S1), allowing a comparison with the structure of the HS gp12093TH057 coree (the CRF01_AE 93TH057 gp120 coree with a single-point H375S mutation) in complex with DMJ-II-121, a compound closely related to BNM-III-170 (PDB ID 4I54). DMJ-II-121 lacks a (methylamino)-methyl addition to position 5 of the indane ring but is identical to BNM-III-170 in all other respects.
Electron density maps of CD4mc in complex with gp120. A 2Fo-Fc electron density map contoured at 1 s shows the density around MCG-IV-210 for the LM+HS gp12093TH057 coree (left) and the LM+HT gp12093TH057 coree (right). MCG-IV-210 is colored gray and gp120 green. Nitrogens are colored blue and oxygens red. Download FIG S3, TIF file, 2.9 MB (2.9MB, tif) .
Copyright © 2020 Prévost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
X-ray crystallography data collection and refinement statistics. Download Table S1, DOCX file, 0.02 MB (17.4KB, docx) .
Copyright © 2020 Prévost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
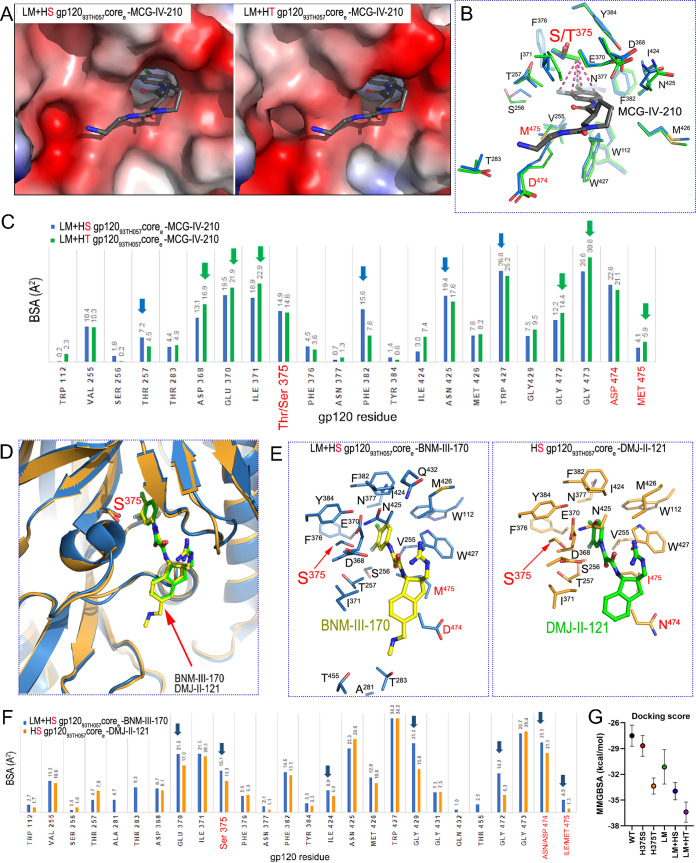
We first sought to understand the exclusive role of residue 375, in the context of the CRF01_AE LM gp120 variant, in influencing the interaction with CD4mc. We analyzed the complex structures of LM+HS and LM+HT gp12093TH057 corees bound to the same MCG-IV-210 compound, which displayed the highest sensitivity to position 375 changes among all CD4mc tested (Fig. 7). As shown in Fig. 8A, MCG-IV-210 bound within the hydrophobic Phe43 cavity of both gp120 mutants, anchoring deeply into the cleft. The buried surface area (BSA) measurements of the complexes were 678 Å2 (243 Å2 from gp120 and 435 Å2 from the compound) and 699 Å2 (252 Å2 from gp120 and 447 Å2 from the compound) for the complexes of LM+HS gp12093TH057 coree–MCG-IV-210 and LM+HT gp12093TH057 coree–MCG-IV-210, respectively, indicating that more surface was buried by the LM+HT variant (Fig. 8C). In addition, while the orientations of MCG-IV-210 in the binding pocket of both complexes were identical, the shapes of the pockets differed. The H375T mutation added one methyl group compared with the H375S mutation, and this addition led to the closing of the Phe43 cavity from the top. The T375 γ-carbon methyl group made a number of additional contacts with the halogenated aromatic ring of the compound, but this did not lead to an increase in the BSA at position 375 due to rearrangement of the Phe43 cavity. In fact, the presence of Thr375 led to a notable decrease in the BSA within the cavity, where residues Thr257 on the left side, Phe382 and Asn425 on the right side, and Trp427 on the bottom made more contact with the compound in the LM+HS mutant. However, residues surrounding the cavity vestibule, including Asp368, Glu370, and Ile371 at the top of the Phe43 cavity, Asn424 on the right side, and Gly472, Gly473, and Met475 at the bottom (as orientated in Fig. 8B), made more contact with MCG-IV-210 and increased the BSA in the LM+HT mutant. The increases in BSA for residues lining the Phe43 cavity may further augment the LM mutant stabilization of layer 3, which may provide an explanation for the added potency for MCG-IV-210 in the LM+HT mutant.
FIG 8.
Structural effects of Phe43 cavity and inner domain changes on CD4mc docking into the Phe43 cavity. (A) Magnified views of the CD4 binding pocket of LM+HS and LM+HT gp12093TH057 core with MCG-IV-210 bound. Complexes are superimposed based on the gp120 core, and electrostatic surfaces are displayed over the gp120 molecule, with blue for electropositive and red for electronegative. The MGC-IV-210 compound is shown as balls and sticks. The LM+HT gp12093TH057 core–MCG-IV-210 complex shown is from PDB ID 6P9N. (B) Details of MCG-IV-210 interactions within the binding pocket. The side chains of gp120 residues that interact with MCG-IV-210 are shown as sticks; LM+HS is depicted in green and LM+HT in blue. MCG-IV-210 is shown with balls and sticks. MCG-IV-210 atoms that interact with the T375 γ-carbon methyl group are shown with dashes connecting to the methyl group. (C) Analysis of the MCG-IV-210 interface for binding to LM+HS and LM+HT gp120. The relative contributions of gp120 residues to compound binding are shown as buried surface area (BSA) data as calculated by PISA. BSA data represent the solvent-accessible surface area of the corresponding residue that is buried upon interface formation. (D) Structural comparison of LM+HS gp12093TH057 coree–BNM-III-170 and HS gp12093TH057 coree–DMJ-II-121 (PDB ID 4I54) complex structures. Complexes are superimposed based on the gp120 core. CD4mc compounds and Ser375 are shown as sticks. (E) Details of binding of BNM-III-170 (left panel) and DMJ-II-121 (right panel) to LM+HS gp12093TH057 coree and HS gp12093TH057 coree, respectively. Residues forming the binding site for the CD4mc compound are shown as sticks (with Gly residues omitted) with LM residues labeled in red. (F) Analysis of the gp120 corees-CD4mc binding interfaces. The relative contributions of gp120 residues to BNM-III-170 and DMJ-II-121 binding are shown by the buried surface area (BSA) as calculated by PISA. BSA represents the solvent-accessible surface area of the corresponding residue that is buried upon interface formation. (G) Docking score based on MMGBSA interaction energies, showing five replica simulations each along with averages, for six BNM-III-170-bound gp120 models.
Next, to better understand the exclusive role of the LM residues in reshaping the Phe43 cavity for effective interaction with CD4mc, we analyzed the complex structures of BNM-III-170 and DMJ-II-121 cocrystalized with gp12093TH057 corees that differed only in the LM residues (Fig. 8D). Structural analyses indicated that the LM+HS gp120 coree–BNM-III-170 complex buried more BSA and was stabilized by more contacts than the HS gp120 coree–DMJ-II-121 complex (Fig. 8E and F). Although some contacts observed in LM+HS gp120 coree–BNM-III-170 complex were not present in HS gp120 coree–DMJ-II-121 complex due to its smaller size [i.e., the hydrogen bond extending from the carbonyl of Gly472 to the (methylamino)methyl addition to BNM-III-170 and contacts extending from the (methylamino)methyl to Ala281, Thr283, and Thr455; Fig. 8F], there was a significant increase in the BSA of the remaining contact residues; these contacted equivalent compound atoms in the BNM-III-170 LM+HS gp120 coree complex {313 Å2 for BNM-III-170 [296 Å2 when the (methylamino)methyl contribution is subtracted] versus 271 Å2 for DMJ-II-121}. These included contacts mediated by the highly conserved Glu370 and Ile424 residues as well as Ser375 itself, all capping the CD4mc in the Phe43 cavity from the top. There were also increased contacts to the 470-to-475 loop region (e.g., residues Gly472, Asp474, and Met475). Interestingly, two of the LM residues, Asp474 and Met475, contributed directly to CD4mc binding and increased BSA. The LM Ile475-to-Met mutation decreased the size of the Phe43 cavity and increased the hydrophobic surface available for CD4mc binding. On the other hand, Asp474 stabilized the interlayer interactions by forming a hydrogen bond to Arg476. This tightening of one side of the Phe43 binding pocket likely also led to the increase in BSA for residues on the opposite side of the pocket, including Ser375 and Gly429 (Fig. 8F). Thus, although only two of the LM residues interacted directly with BNM-III-170, their combination had an additive effect on binding and resulted in an overall increase in the complex BSA and compound affinity.
Furthermore, we conducted in silico analysis to predict the interaction energies present upon docking of CD4mc BNM-III-170 with our different gp120 variants. All-atom, explicit-solvent molecular dynamics (MD) simulations of BNM-III-170-bound complexes were conducted for the WT, H375S, H375T, LM, LM+HS, and LM+HT versions of gp120 coree. Average protein-ligand interaction energies were computed from these simulations under the generalized Born surface area (GBSA) implicit solvent model. The average relative interaction energies ranked from least to most favorable in the order WT > H375S > LM > H375T > LM+HS > LM+HT, as shown in Fig. 8G. This ordering is consistent with the ordering in half-maximal inhibitory concentration (IC50) values indicated in Fig. 3D, suggesting that enthalpic interactions are of high importance in determining how these residues change and how they reshape the CD4-binding site (CD4BS) to affect the activity of CD4mc.
Phe43 cavity and inner domain substitutions shape the highly conserved CD4 binding site.
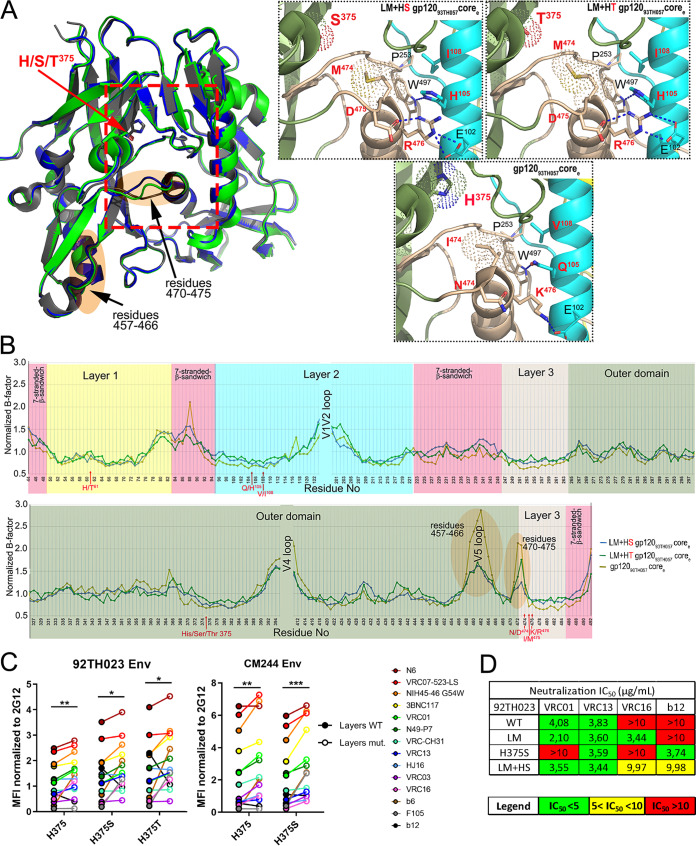
The CD4-binding site (CD4BS) region of Env represents a highly complex quaternary arrangement of five different loops that meet to form this highly conserved structure. The following three loops converge to form the Phe43 cavity: the CD4-binding loop (residues 365 to 371), which makes critical contacts with CD4 residues F43 and R59 (1); the β20-β21 loop (residues 424 to 432), which acts as a conformational regulatory switch between the inner domain and the outer domain of gp120 (62); and the outer domain exit loop (residues 470 to 475), linking the outer domain to inner domain layer 3. Two other loops in the periphery of the Phe43 cavity were shown to be implicated in CD4 binding: loop D (residues 275 to 283) and the V5 loop (residues 457 to 468) (1). On the basis of structural analysis presented in Fig. 8, the LM+HS/T changes appear to have increased the interaction of CD4mc with the Phe43 cavity by restructuring the CD4BS, most particularly the outer domain exit loop. To better understand the molecular basis of this phenomenon, we solved crystal structures of gp12093TH057 corees containing the LM mutations in combination with H375S (LM+HS) or H375T (LM+HT) in their unliganded state (Table S1). Unliganded structures of the LM+HS and LM+HT cores were solved at 2.2-Å and 2.5-Å resolution, respectively. The structures of LM+HS and LM+HT gp12093TH057 corees compared to the wild-type gp12093TH057 coree (PDB code 3TGT [49]), which carries a His at position 375, are shown in Fig. 9.
FIG 9.
Effect of gp120 layer mutations on the highly conserved CD4 binding site. (A) Structures of LM+HS and LM+HT gp12093TH057 unliganded corees are superimposed onto wild-type gp12093TH057 coree (PDB code: 3TGT). Residue 375 is shown as sticks, and the gp120 regions proximal to the CD4 binding site that shows the lowest B-factors in the LM core mutants compared to the unmutated core are highlighted in orange. The magnified views (right panel) show the network of interactions in each core mediated by LM+HS/T residues. The hydrogen bonds are shown as blue dashes. LM+HS/T residues are labeled in red. (B) Plot of the normalized B-factors for the main chain atoms of residues of coree structures. The rigidified region is highlighted in orange. (C) Cell surface staining of 293T cells transfected with CRF01_AE Env expressors (92TH023 and CM244 isolates [WT or their mutated counterparts]) using a panel of CD4-binding site antibodies (CD4BS Abs). Shown are the mean fluorescence intensities (MFI) normalized to 2G12 MFI obtained in the transfected (GFP+) population for staining obtained in at least 3 independent experiments. All MFI data were normalized to 2G12 MFI for each Env mutant. Error bars indicate means ± SEM. Statistical significance was tested using a paired t test or a Wilcoxon signed-rank test based on statistical normality (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (D) Recombinant HIV-1 strains expressing luciferase and bearing wild-type or mutant CRF01_AE Envs (92TH023 and CM244 isolates) were normalized by reverse transcriptase activity. Normalized amounts of viruses were incubated with serial dilutions of VRC01, VRC16, VRC13, or b12 at 37°C for 1 h prior to infection of Cf2Th-CD4/CCR5 cells. Neutralization half-maximal inhibitory concentration (IC50) data are summarized.
There is close similarity between the structures of the LM+HS and LM+HT corees. Indeed, alignments showed that they have a root mean square deviation (RMSD) of 0.43 Å for their main chain atoms. Alignments to the wild-type gp12093TH057 coree, resulted in RMSD values of 0.45 Å and 0.54 Å for the LM+HS and LM+HT mutants, respectively, indicating a larger dissimilarity between the LM+HT and wild-type coree structures. Interestingly, a closer look into interactions formed by mutated residues in the LM+HS and LM+HT corees (Fig. 9A, magnified view) reveals a new network of interactions mediated by Asp474 and Arg476 that stabilize the inner domain mobile layer 2 and 3 interface in the CD4-bound conformation (51, 63, 64). Asp474 in combination with Glu102 in layer 2 formed a network of hydrogen bonds with Arg476 in both LM mutants that bridged the two layers. This network of hydrogen bonds is absent from other clade gp120 coree structures, with the exception of the clade B YU2 gp120 (PDB ID 3TGQ). In wild-type CRF01_AE gp120, Lys476 formed a hydrogen bond with Glu102, but without the added stability made possible by the Arg476 guanidinium group, leaving Asn474 out of the interface. Other LM mutant changes that potentially add to the interface stability are the Gln105-to-His mutation, which changes the character of the Trp479 amide hydrogen bond, and the Val108-to-Ile mutation, which increases the hydrophobic surface available for interaction with Pro253. CRF01_AE 93TH057 gp120 has a histidine at residue 375, in contrast to the more common Ser or Thr, which only partially fills the Phe43 cavity, leaving the space required for CD4mc compound binding. The H375S/T mutation most likely induces a loss in stability of the CD4 binding pocket in the absence of CD4 or CD4mc that in LM variants is partially compensated for by the Ile475-to-Met mutation. Both Ile475 in the wild-type gp120 and Met475 in the LM mutant pack against Trp479, which links the Phe43 pocket to the layer 3-layer 2 interface through the Trp479 amide bond to Gln105 (wild type) or His105 (LM). The bulkier side chain of Met475 provides more hydrophobic surface to the network, stabilizing and reshaping the Phe43 cavity.
The added stability of the LM mutations can be visualized by the crystallographic temperature or B-factors which describe the spread of electron density attributed to each atom (Fig. 9B). Outside the V1/V2 and V4 loop regions, which differ greatly between structures, one region formed by residues 470 to 475 stands out in the normalized B-factor plot (65). The levels of main chain atom B-factors corresponding to the residues in this region are much lower for the LM+HS and LM+HT mutants than for wild-type Env, implying that there is a greater degree of structural stability in this region in the LM mutants. It was shown previously that residues of the outer domain exit loop contribute to CD4mc binding, as also seen in the unliganded LM+HS and LM+HT corees (37, 45, 48). This raises the possibility that the observed rigidification constitutes an element that reshapes the Phe43 cavity, leading to preferential binding of CD4 or CD4mc; conversely, binding of CD4 or CD4mc might stabilize layer 3, which can lead to downstream conformational changes in the gp120 inner domain. Interestingly, another region of the CD4BS was found to be significantly rigidified in the presence of the LM+HS/T mutations: the V5 loop region (residues 457 to 468). This region has previously been described as being implicated in CD4 attachment but also as being recognized by most known broadly neutralizing Abs targeting the CD4BS (e.g., VRC01). Notably, alterations in the V5 region have been associated with resistance to members of the VRC01 class of Abs (66–71) and b12 (72), consistent with a potential effect of V5 loop rigidification on gp120 recognition by CD4BS Abs. To validate our model, we evaluated the ability of a panel of CD4BS Abs to bind to CRF01_AE Envs harboring one of several different residues at position 375 (H375, S375, and T375) in combination with the LM mutations or not. Consistent with the CD4BS restructuring shown in Fig. 9A, the presence of the LM mutations was shown to increase the binding of all CD4BS Abs, except b12, regardless of the nature of the residue at position 375 in both CRF01_AE strains tested (Fig. 9C). CD4BS recognition of Env-expressing cells correlated with the neutralization profile of these mutants (Fig. 9D).
Finally, since changes in the gp120 inner domain layers in combination with changes at position 375 have a global effect on the CD4BS, we tested the sensitivity of our panel of CRF01_AE Env mutants to another class of Env antagonists targeting the CD4BS: cyclic peptide triazoles (cPTs) (73, 74). Again, the introduction of the LM mutations in combination with a small residue at position 375 (LM+HS and LM+HT) in a CRF01_AE Env was found to dramatically increase the sensitivity to neutralization by all cPTs tested (AAR029N2 and AAR029N3) (Fig. S4). However, the presence of a bulky residue at position 375 (H375) was found to abrogate this effect and conferred total resistance to the members of this class of compound (Fig. S4B to D). In summary, supported by structural data and functional assays using a wide variety of CD4BS probes, our data show that residue 375 and its inner domain coevolving residues act together to shape the highly conserved CD4BS.
Phe43 cavity and inner domain changes enhance the sensitivity of CRF01_AE strains to neutralization by cPTs. (A) Chemical structure of the different cPTs tested. (B to D) Recombinant HIV-1 strains expressing luciferase and bearing wild-type or mutant CRF01_AE Envs (92TH023 isolate) were normalized by reverse transcriptase activity. Normalized amounts of viruses were incubated with serial dilutions of the cyclic peptide triazoles (cPTs) AAR029N2 (B) and AAR029N3 (C) at 37°C for 1 h prior to infection of Cf2Th-CD4/CCR5 cells. Infectivity at each dilution of cPTs tested is shown as the percentage of infection without cPTs for each particular mutant. Quadruplicate samples were analyzed in each experiment. Data shown are the means of results obtained in at least 3 independent experiments. The error bars represent the standard deviations. Neutralization half-maximal inhibitory concentrations (IC50) are summarized in panel D. Download FIG S4, TIF file, 0.3 MB (282.4KB, tif) .
Copyright © 2020 Prévost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Previous studies have indicated that the gp120 inner domain plays a role in modulating the transition of Env from its unbound to its CD4-bound conformation (13–15, 75, 76). Here, we add a detailed structural understanding of this mechanism by identifying six gp120 inner domain residues able to shape the Phe43 cavity and the CD4 binding site. Collectively named layer mutants (LM), these residues determine the efficiency with which HIV-1 Envs negotiate transitions from the pretriggered state 1 conformation to CD4-bound conformations (states 2 and 3). Structures of trimeric Env stabilized in the native state 1 conformation might be required for observation of the conformational rearrangement of the gp120 inner domain layers upon CD4 or CD4mc binding. The efficiency with which a given HIV-1 variant undergoes these conformational changes influences virus susceptibility to a number of antiviral agents directed against the CD4-binding region of gp120, including CD4mc, CD4 miniproteins, cPTs, and different CD4BS antibodies. Our report documents the significant impact of the LM residues on the susceptibility of HIV-1 strain variants to this major class of virus entry inhibitors and opens the door to the rational design of agents with improved antiviral potency and breadth.
The LM residues also modulate the susceptibility of infected cells to ADCC mediated by HIV+ sera. Small-molecule CD4mc have been shown to enhance dramatically the ADCC-mediating potency of the antibodies present at high titers in the sera of almost all HIV-1-infected individuals (33). CD4mc are under investigation as a means to use ADCC to effect a functional “cure” of HIV-1 infection, as an adjunct to highly active antiretroviral therapy. Knowledge of the identity of the LM residues in the HIV-1 strains present in infected individuals could assist in the identification of subjects most likely to benefit from available CD4mc. The structural information on LM residues provided by our study can assist the design of CD4mc tailored to be effective against particular HIV-1 strains.
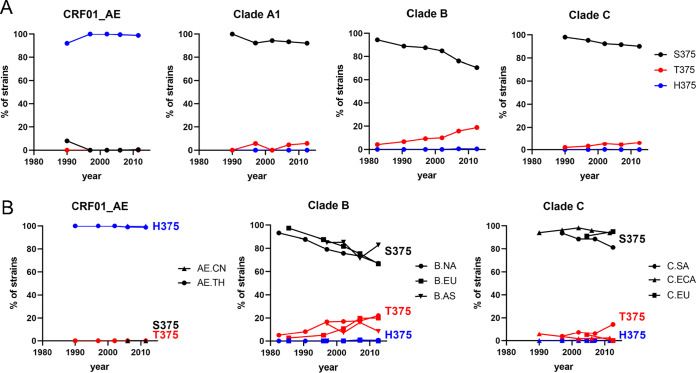
Many epitopes of Env have changed at a population level during the AIDS pandemic (77). Notably, a significant decline in sensitivity of clade B circulating strains to CD4BS antibodies was observed. We examined the historical changes in residue occupancy at position 375 in diverse clades and distinct geographic regions. Among CRF01_AE strains, H375 remained highly conserved during the pandemic (Fig. 10A). In contrast, striking changes occurred in other group M major clades, most importantly in clade B, where a gradual but constant loss of S375 occurred, with S375 progressively replaced by T375 (Fig. 10A). The different patterns observed in the HIV-1 major clades and CRF01_AE, which follow similar patterns in distinct geographic regions (Fig. 10B), likely reflect differences in the constraints applied to the Phe43 cavity in the unique context of their Envs. They suggest an ongoing evolution of theCD4BS, which might need to be taken into account in developing CD4mc or immunogens designed to elicit CD4BS Abs. With respect to the issue of why T375 seems especially able to induce susceptibility to CD4mc irrespective of the LM, one clue from the structures presented here offers a possible explanation: space in the Phe43 cavity near S375 is strongly occupied by a density that may represent water or a crystallization solute, and this density is largely lacking for T375; these observations suggest higher susceptibility for T375 because it has fewer solvent molecules to eject in permitting CD4mc binding.
FIG 10.
Historical changes in amino acid sequence at position 375 in diverse clades and distinct geographic regions. Env sequences from samples collected worldwide between 1979 and 2015 were examined. Totals of 335, 1,942, 1,248, and 543 Envs were used, from clades A1, B, C, and CRF01_AE, respectively. (A) Changes in frequencies of residues at position 375 in different clades. Values represent percentages of isolates from each 5-to-7-year period that contained the indicated residue. (B) Changes in frequencies of residues at position 375 in the indicated geographic regions. The residues shown in panels A and B account in most cases for more than 95% of all circulating strains. Sequences were analyzed phylogenetically to include a single Env from each patient (see Materials and Methods). For accuracy, only data from time periods with more than 25 patients are included for each clade and region. The following abbreviations are used: for clade B, NA, North America; EU, Europe; AS, Asia; for clade C, SA, Southern Africa (South Africa and Botswana); ECA, Eastern and Central Africa; IN/NP, India and Nepal; for CRF01_AE, CN, China; TH, Thailand.
Taking the data together, here we show that residue 375 and its inner domain coevolving residues act together to shape the highly conserved CD4BS. This new structural understanding will assist ongoing efforts to improve CD4mc and other agents directed against the CD4BS and to design immunogens aimed at eliciting CD4BS antibodies.
MATERIALS AND METHODS
Ethics statement.
Written informed consent was obtained from all study participants (the Montreal Primary HIV Infection Cohort [78, 79] and the Canadian Cohort of HIV Infected Slow Progressors [80–82]), and research adhered to the ethical guidelines of CRCHUM and was reviewed and approved by the CRCHUM institutional review board (ethics committee, approval number CE 16.164–CA). Research adhered to the standards indicated by the Declaration of Helsinki. All participants were adults and provided informed written consent prior to enrollment in accordance with Institutional Review Board approval.
Sequence analysis.
The Logo plots (83) for HIV were made using the Analyze Align tool at the HIV database and are based on the WebLogo 3 program (https://www.hiv.lanl.gov/content/sequence/ANALYZEALIGN/analyze_align.html) and the HIV-1 database global curated and filtered 2017 alignment published circa June 2018, including 1 HIV-1 Env protein sequence per person from 5,471 individuals. The relative height of each letter within individual stack represents the frequency of the indicated amino acid at that position. The numbering of all the Env amino acid sequences is based on the prototypic HXBc2 strain of HIV-1, where 1 is the initial methionine (84).
Analysis of historical changes in amino acid sequence.
HIV-1 env sequences were obtained from the Los Alamos National Lab (LANL) database (https://www.hiv.lanl.gov). Nonfunctional Envs and sequences with nucleotide ambiguities were removed. Nucleotide sequences were aligned using a hidden Markov model with HMMER3 software (85), and phylogenetic relationships between sequences were inferred by the maximum-likelihood method using PhyML3 (86). A single env gene from each patient and a single sequence from known transmission pairs were used. In addition, a minimal distance of 0.03 nucleotide substitutions per site was applied as a cutoff for selection. Sequences were archived and residue frequencies at all Env positions in each population were determined using software developed in-house, as previously described (77).
Cell lines and isolation of primary cells.
HEK293T human embryonic kidney cells and Cf2Th canine thymocytes (obtained from ATCC) were grown as previously described (14). Cf2Th cells stably expressing human CD4 and CCR5 (Cf2Th-CD4/CCR5) (87) were grown in medium supplemented with 0.4 mg/ml of G418 (Invitrogen) and 0.15 mg/ml of hygromycin B (Roche Diagnostics). Primary human peripheral blood mononuclear cells (PBMCs) and CD4+ T cells were isolated, activated, and cultured as previously described (23, 33). Briefly, PBMCs were obtained by leukapheresis and CD4+ T lymphocytes were purified from resting PBMCs by negative selection using immunomagnetic beads per the instructions of the manufacturer (StemCell Technologies, Vancouver, BC, Canada) and were activated with phytohemagglutinin-L (10 μg/ml) for 48 h and then maintained in RPMI 1640 complete medium supplemented with recombinant interleukin-2 (rIL-2) (100 U/ml).
Plasmids and proviral constructs.
The plasmids expressing the CRF01_AE Envs HIV-192TH023 and HIV-1CM244 were previously reported (41, 88). Plasmid pSVIIIenv expressing the full-length HIV-1YU2 Env and Tat-expressing plasmid pLTR-Tat were previously described (14). The sequence of full-length clade B HIV-1JRFL (21) and clade A HIV-1BG505 Envs (89) were codon optimized (GenScript) and cloned into expression plasmid pcDNA3.1(-) (Invitrogen). An asparagine residue was introduced at position 332 (N332) to allow recognition of 2G12 antibody. Vesicular stomatitis virus G (VSV-G)-encoding plasmid pSVCMV-IN-VSV-G was previously described (90). For crystallographic studies, the plasmid used to express gp120 extended core (coree) from CRF01_AE strain 93TH057 (gp120 lacking the N and C termini and variable loops 1, 2, and 3) was previously described (49).The transmitted/founder (TF) CRF01_AE 40061 full viral genome was retrieved by single-genome amplification and cloned to generate full-length infectious molecular clone (IMC) as previously reported (91). TF and chronic IMCs of patients CH40, CH58, CH77, CH167, CH185, CH198, CH236, CH470, CH505, CH850, RHGA, and STCO were inferred, constructed, and biologically characterized as previously described (92–98). The JRFL IMC was also previously described (99).Mutations were introduced individually or in combination into the different Env expressors or IMCs using the QuikChange II XL site-directed mutagenesis protocol (Stratagene). The presence of the desired mutations was determined by automated DNA sequencing. The numbering of all the Env amino acid sequence is based on the prototypic HXBc2 strain of HIV-1, where 1 is the initial methionine (84).
Viral production and infections.
To achieve similar levels of infection in primary CD4+ T cells among the different IMCs tested, VSV-G-pseudotyped HIV-1 was produced and titrated as previously described (24). Viruses were then used to infect activated primary CD4 T cells from healthy HIV-1-negative donors by spin infection at 800 × g for 1 h in 96-well plates at 25°C. For the viral neutralization assay, Cf2Th-CD4/CCR5 cells were infected with single-round luciferase-expressing HIV-1 (14). Briefly, 293T cells were transfected by the calcium phosphate method with the proviral vector pNL4.3(Env-)Luc and a plasmid expressing wild-type or mutant HIV-1 Env at a ratio of 2:1. Two days after transfection, the cell supernatants were harvested. The reverse transcriptase activities of all virus preparations were measured as described previously (100). Each virus preparation was frozen and stored in aliquots at –80°C until use.
Antibodies and sera.
The following Abs were used to assess cell surface Env staining: A32, 17b, 19b, F240, PG16, N6, NIH45-46 G54W, VRC01, VRC03, HJ16, VRC-CH31, F105 (NIH AIDS Reagent Program), PGT151, PGT128 (IAVI), 3BNC117 (kindly provided by M. Nussenzweig, Rockefeller Institute, New York, NY), b6, b12 (kindly provided by D. Burton, The Scripps Research Institute, La Jolla, CA), VRC07-523-LS, VRC13, VRC16 (kindly provided by J. Mascola, Vaccine Research Center, Bethesda, MD), GE2.JG8 (kindly provided by G. Karlsson Hedestam, Karolinska Institutet, Sweden), and N49P7 (kindly provided by M. Sajadi, University of Maryland, Baltimore, MD). The anti-CD4 OKT4 monoclonal antibody (recognizing the D3 domain of CD4) (eBioscience) was used to measure the binding of sCD4 to cell surface Env. Goat anti-mouse and anti-human antibodies precoupled to Alexa Fluor 647 (Invitrogen) were used as secondary antibodies in flow cytometry experiments. Sera from HIV-infected individuals were collected, heat inactivated, and conserved at −80°C until use. A random number generator (GraphPad; QuickCalcs, San Diego, CA, USA) was used to randomly select a number of sera for each experiment.
CD4 mimetics and other small molecules.
Soluble CD4 (sCD4) and the miniprotein M48U1 were produced and purified as previously described (14, 101). The CD4 mimetic compounds (CD4mc) NBD-556, JRC-II-191, DMJ-II-121, JP-III-48, BNM-III-170, BNM-IV-147, BNM-IV-197, MCG-IV-210, SMK-II-48, AEG-I-249, AEG-I-259, and MCG-III-051 were developed and synthesized as described previously (37, 45–47, 52, 61, 102) or as described in the “Chemical synthesis” section below. The CD4mc were analyzed, dissolved in dimethyl sulfoxide (DMSO) at a stock concentration of 10 mM, aliquoted, and stored at −80°C. Cyclic peptide triazoles (cPTs) AAR029N2 and AAR029N3 were synthesized, purified (>95% homogeneity by reverse-phase HPLC), and structurally validated as previously described (73, 74). The cPTs were analyzed, dissolved in DMSO at a stock concentration of 10 mM, aliquoted, and stored at −80°C. Each compound was then diluted to the indicated concentrations in phosphate-buffered saline (PBS) for cell surface staining, in RPMI 1640 complete medium for ADCC assays, or in complete Dulbecco’s modified Eagle’s medium (DMEM) for viral neutralization assays.
Viral neutralization assay.
Cf2Th-CD4/CCR5 target cells were seeded at a density of 5 × 103 cells/well in 96-well luminometer-compatible tissue culture plates (Perkin Elmer) 24 h before infection. Luciferase-expressing recombinant viruses (10,000 reverse transcriptase units) in a final volume of 100 μl were incubated with the indicated amounts of different proteins, compounds, or antibodies for 1h at 37°C and were then added to the target cells followed by incubation for 48 h at 37°C; the medium was then removed from each well, and the cells were lysed by the addition of 30 μl of passive lysis buffer (Promega) followed by three freeze-thaw cycles. An LB 941 TriStar luminometer (Berthold Technologies) was used to measure the luciferase activity of each well after the addition of 100 μl of luciferin buffer (15 mM MgSO4, 15 mM KPO4 [pH 7.8], 1 mM ATP, and 1 mM dithiothreitol) and 50 μl of 1 mM d-luciferin potassium salt (Prolume). The neutralization half-maximal inhibitory concentration (IC50) represents the amount of protein, compound, or antibody needed to inhibit 50% of the infection of Cf2Th-CD4/CCR5 cells by recombinant luciferase-expressing HIV-1 bearing the indicated Env.
Flow cytometry analysis of cell surface staining.
Cell surface staining of infected cells was performed as previously described (24, 33). Binding of cell surface HIV-1 Env by sera (1:1,000 dilution) or anti-Env MAbs (5 μg/ml) was performed at 48 h postinfection in the presence of CD4mc or of an equal amount of the vehicle (DMSO). Infected cells were stained intracellularly for HIV-1 p24, using a Cytofix/Cytoperm fixation/permeabilization kit (BD Biosciences, Mississauga, ON, Canada) and fluorescent anti-p24 MAb (phycoerythrin [PE]-conjugated anti-p24, clone KC57; Beckman Coulter/Immunotech). The percentage of infected cells (p24+) was determined by gating the living cell population on the basis of viability dye staining (Aquavivid; Thermo Fisher Scientific). Samples were acquired on an LSR II cytometer (BD Biosciences), and data analysis was performed using FlowJo vX.0.7 (Tree Star, Ashland, OR, USA).
Cell surface staining of Env-expressing 293T cells was performed as previously described (103). Briefly, 2 × 106 cells were transfected with 7 μg of Env expressor and 1 μg of a green fluorescent protein (GFP) expressor (pIRES-GFP) with the calcium-phosphate method. When the pSVIII Env expressor was used, it was cotransfected with 0.25 μg of a Tat-expressing plasmid. At 48 h posttransfection, 293T cells were stained with anti-Env antibodies (5 μg/ml). Alternatively, to evaluate sCD4 binding to the different cell surface Envs, transfected 293T cells were incubated with sCD4 (5 μg/ml), followed by staining performed with the monoclonal anti-CD4 OKT4 antibody (0.5 μg/ml). In experiments using transfected cells, all mean fluorescent intensities (MFI) were normalized to the MFI of 2G12 for each Env mutant.
FACS-based ADCC assay.
Measurement of ADCC using the fluorescence-activated cell sorter (FACS)-based assay was performed at 48 h postinfection as previously described. Briefly, infected primary CD4+ T cells were stained with Aquavivid viability dye and cell proliferation dye (eFluor670; eBioscience) and used as target cells. Autologous PBMC effector cells, stained with another cellular marker (cell proliferation dye eFluor450; eBioscience), were added at an effector/target ratio of 10:1 in 96-well V-bottom plates (Corning, Corning, NY). A 1:1,000 final dilution of sera was added to appropriate wells, and the cells were incubated for 15 min at room temperature. The plates were subsequently centrifuged for 1 min at 300 × g and incubated at 37°C and 5% CO2 for 5 h before being fixed in a 2% PBS–formaldehyde solution. Samples were acquired on an LSR II cytometer (BD Biosciences), and data analysis was performed using FlowJo vX.0.7 (Tree Star). The percentage of ADCC resulting from gating performed on infected lived target cells was calculated with the following formula: (percentage of p24+ cells in targets plus effectors) − (percentage of p24+ cells in targets plus effectors plus sera)/(percentage of p24+ cells in targets).
Protein expression and purification.
Plasmids encoding the layer mutant gp120 extended core (coree) proteins, HIV-193TH057 gp120 coree LM+HS and LM+HT, were transfected into GnT1− cells using Xtremegene transfection reagent (Sigma-Aldrich) per the manufacturer’s instructions. Following 7 days of culture growth at 37°C and 8% CO2, cells were pelleted by centrifugation and cell supernatant was filtered. Expressed gp120 was purified by passage over a 17b affinity column made by cross-linking MAb 17b to protein A (Pierce protein A IgG plus orientation kit; Thermo Fisher). gp120 was eluted with 0.1 M glycine (pH 3.0) into tubes containing 1/10 the final volume of 1 M Tris-HCl (pH 8.5) to raise the pH. The protein was then deglycosylated with 10 units/μg of Endo Hf (New England Biolabs) overnight at 37°C in a mixture of deglycosylation buffer, 50 mM sodium acetate (pH 6.0), and 350 mM sodium chloride. Endo Hf was removed by passage over an amylose resin column, and the protein was further purified by gel filtration chromatography on a Superdex 200 16/60 column (GE Healthcare, Piscataway, NJ) equilibrated with 5 mM Tris-HCl (pH 7.2) and 150 mM sodium chloride. The protein was concentrated to approximately 5 mg/ml for use in crystallization trials.
X-ray crystallography.
Deglycosylated HIV-193TH057 gp120 coree LM+HS or LM+HT (5 mg/ml) was crystalized by the hanging drop method in a mixture containing 5 to 10% polyethylene glycol (PEG) 1500, 6% PEG 400 (LM+HT), and 0.1 M HEPES (pH 7.5) or 5 to 10% PEG 3350, 6% PEG 400, and 0.1 M HEPES (pH 7.5) (LM+HS). CD4 mimetics were solubilized with DMSO at a concentration of 10 mM and diluted with crystallization buffer to 100 nM prior to use in crystal soaks. When CD4mc was added to the crystal, an equal volume of the 100 nM mimetic in crystallization buffer was added to the hanging drop and the crystals were allowed to soak for 4 h before being frozen. Crystals were flash frozen in liquid nitrogen after a brief soak in crystallization buffer containing 18% MPD (2-methyl-2,4-pentanediol) for cryoprotection and a 50 nM concentration of CD4mc when CD4mc was added.
Data collection, structure solution, and refinement.
Data were collected on National Synchrotron Light Source II (NSLS II) highly automated macromolecular crystallography (AMX) beamline 17-ID-1 on an Eiger 9M detector system or on Stanford Synchrotron Radiation Light Source (SSRL) beamline 12-2 on a Dectris Pilatus 6M detector system. Data were integrated and processed with MOSFLM and SCALA from the CCP4 suite (104) or HKL2000 (105). Crystals were orthorhombic, belonging to space group P212121, with cell dimensions of a = 64.0 to 66.8 Å, b = 65.6 to 67.5 Å, and c = 86.2 to 87.8 Å, with diffraction to 2.2 to 2.65 Å. Structures were solved by molecular replacement with the program PHASER from the CCP4 suite using PDB ID 3TGT as a starting model. Model building was done using the program COOT (106). Refinement was done using the program REFMAC from the CCP4 suite or PHENIX (65).
Structure validation and analysis.
The quality of the final refined models was monitored using the program MolProbity (107). Structural alignments were performed using the Dali server and the program LSQKAB from the CCP4 suite (104) The PISA webserver was used to determine contact surfaces and residues. All illustrations were prepared with the PyMol molecular graphic suite (DeLano Scientific, San Carlos, CA, USA). The Ramachandran plot was obtained by the use of the validation program “MolProbity” and shows that 92.8 to 97.0% of the total amino acids are in the most favored region, 3 to 6.6% in the generously allowed region, and 0 to 0.6% in the disallowed region, depending on the structure, with better values corresponding to the higher-resolution structures. Complete data collection and refinement statistics can be found in Table S1.
In silico analysis.
Starting with the H375S clade A/E HIV-1 core gp120 monomer in PDB entry 4H8W (108), we generated via residue mutation the WT, H375S, H375T, LM, LM+HS, and LM+HT variants and docked BNM-III-170 to each via alignment to the 5F4P PDB entry (45). For each variant, five independent fully TIP3P-solvated MD systems were generated and run for 500 ns each using NAMD 2.13 with the CHARM36 force field, with configurations saved every 0.1 ns. For each system, molecular mechanics-generalized Born surface area (MMGBSA) interaction energy data were computed by reprocessing trajectories through NAMD and computing protein-only, ligand-only, and protein-plus-ligand potential energies under the generalized Born implicit solvent (GBIS) model. The GBIS parameters included a solvent dielectric constant of 74.69, a total molar ion concentration of 0.3, a screening cutoff of 14 Å, and surface tension of 0.0072 kcal/mol/Å2.
Statistical analysis.
Statistics were analyzed using GraphPad Prism version 6.01 (GraphPad, San Diego, CA, USA). Every data set was tested for statistical normality, and this information was used to apply the appropriate (parametric or nonparametric) statistical test. P values of <0.05 were considered significant, and significance values are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Chemical synthesis.
For details about chemical synthesis, see Text S1 in the supplemental material.
Details about chemical synthesis. Download Text S1, DOCX file, 0.8 MB (809.9KB, docx) .
Copyright © 2020 Prévost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We thank the staff members of the CRCHUM BSL3 and Flow Cytometry Platforms for technical assistance, Mario Legault from the FRQS AIDS and Infectious Diseases network for cohort coordination and clinical samples, Michel Nussenzweig for 3BNC117, and Dennis Burton for infectious molecular clone JRFL.
This work was supported by CIHR foundation grant 352417 to A.F. Support for this work was also provided by NIH R01 to A.F. and M.P. (AI129769) and J.S. (AI124982 and AI145547) and by NIAID R01 to M.P. (AI116274). This study was also supported by NIH R01-GM56550 to A.B.S. and by grant P01-GM56550/AI150741 to A.F., A.B.S., I.C., J.S., and C.F.A. A.F. is the recipient of a Canada Research Chair on Retroviral Entry RCHS0235. J.P. is the recipient of a CIHR doctoral fellowship. C.H. was supported by a Development Grant from the University of Iowa (MI-2019). F.K. is supported by grants from the DFG (CRC 1279 and SPP 1923). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The views expressed in this presentation are those of the authors and do not reflect the official policy or position of the Uniformed Services University, U.S. Army, the Department of Defense, or the U.S. Government.
J.P. and A.F. conceived the study. J.P., M.P., and A.F. designed experimental approaches. J.P., W.D.T., H.M., R.T.S., N.M., D.Z., N.V., C.H., H.H., C.F.A., J.S., M.P., and A.F. performed, analyzed, and interpreted the experiments. G.G.-L., A.E.G., M.C.G., S.K., B.T.M., A.L.C., A.A., I.C., F.K., B.H.H., and A.B.S. supplied novel reagents. J.P., J.S., M.P., and A.F. wrote the paper. All of us read, edited, and approved the final manuscript.
We declare that we have no competing interests.
Footnotes
Citation Prévost J, Tolbert WD, Medjahed H, Sherburn RT, Madani N, Zoubchenok D, Gendron-Lepage G, Gaffney AE, Grenier MC, Kirk S, Vergara N, Han C, Mann BT, Chénine AL, Ahmed A, Chaiken I, Kirchhoff F, Hahn BH, Haim H, Abrams CF, Smith AB, III, Sodroski J, Pazgier M, Finzi A. 2020. The HIV-1 Env gp120 inner domain shapes the Phe43 cavity and the CD4 binding site. mBio 11:e00280-20. https://doi.org/10.1128/mBio.00280-20.
Contributor Information
Tongqing Zhou, Vaccine Research Center, National Institute of Allergy and Infectious Diseases.
Thomas E. Smithgall, University of Pittsburgh School of Medicine.
REFERENCES
- 1.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madani N, Perdigoto AL, Srinivasan K, Cox JM, Chruma JJ, LaLonde J, Head M, Smith AB III, Sodroski JG. 2004. Localized changes in the gp120 envelope glycoprotein confer resistance to human immunodeficiency virus entry inhibitors BMS-806 and #155. J Virol 78:3742–3752. doi: 10.1128/JVI.78.7.3742-3752.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 4.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 5.Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, Peiper SC, Parmentier M, Collman RG, Doms RW. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 6.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 7.Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 8.Feng Y, Broder CC, Kennedy PE, Berger EA. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 9.Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 10.Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 11.Lu M, Blacklow SC, Kim PS. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol 2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 12.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 13.Desormeaux A, Coutu M, Medjahed H, Pacheco B, Herschhorn A, Gu C, Xiang SH, Mao Y, Sodroski J, Finzi A. 2013. The highly conserved layer-3 component of the HIV-1 gp120 inner domain is critical for CD4-required conformational transitions. J Virol 87:2549–2562. doi: 10.1128/JVI.03104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finzi A, Xiang SH, Pacheco B, Wang L, Haight J, Kassa A, Danek B, Pancera M, Kwong PD, Sodroski J. 2010. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Mol Cell 37:656–667. doi: 10.1016/j.molcel.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding S, Tolbert WD, Prevost J, Pacheco B, Coutu M, Debbeche O, Xiang SH, Pazgier M, Finzi A. 12 September 2016, posting date A highly conserved gp120 inner domain residue modulates Env conformation and trimer stability. J Virol doi: 10.1128/JVI.01068-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finzi A, Pacheco B, Xiang SH, Pancera M, Herschhorn A, Wang L, Zeng X, Desormeaux A, Kwong PD, Sodroski J. 2012. Lineage-specific differences between human and simian immunodeficiency virus regulation of gp120 trimer association and CD4 binding. J Virol 86:8974–8986. doi: 10.1128/JVI.01076-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding S, Medjahed H, Prevost J, Coutu M, Xiang SH, Finzi A. 2016. Lineage-specific differences between the gp120 inner domain layer 3 of human immunodeficiency virus and that of simian immunodeficiency virus. J Virol 90:10065–10073. doi: 10.1128/JVI.01215-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu M, Ma X, Castillo-Menendez LR, Gorman J, Alsahafi N, Ermel U, Terry DS, Chambers M, Peng D, Zhang B, Zhou T, Reichard N, Wang K, Grover JR, Carman BP, Gardner MR, Nikic-Spiegel I, Sugawara A, Arthos J, Lemke EA, Smith AB III, Farzan M, Abrams C, Munro JB, McDermott AB, Finzi A, Kwong PD, Blanchard SC, Sodroski JG, Mothes W. 2019. Associating HIV-1 envelope glycoprotein structures with states on the virus observed by smFRET. Nature 568:415–419. doi: 10.1038/s41586-019-1101-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB III, Kwong PD, Blanchard SC, Mothes W. 2014. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao Y, Wang L, Gu C, Herschhorn A, Xiang SH, Haim H, Yang X, Sodroski J. 2012. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nat Struct Mol Biol 19:893–899. doi: 10.1038/nsmb.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veillette M, Coutu M, Richard J, Batraville LA, Desormeaux A, Roger M, Finzi A. 14 September 2014, posting date Conformational evaluation of HIV-1 trimeric envelope glycoproteins using a cell-based ELISA assay. J Vis Exp doi: 10.3791/51995:51995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veillette M, Desormeaux A, Medjahed H, Gharsallah NE, Coutu M, Baalwa J, Guan Y, Lewis G, Ferrari G, Hahn BH, Haynes BF, Robinson JE, Kaufmann DE, Bonsignori M, Sodroski J, Finzi A. 2014. Interaction with cellular CD4 exposes HIV-1 envelope epitopes targeted by antibody-dependent cell-mediated cytotoxicity. J Virol 88:2633–2644. doi: 10.1128/JVI.03230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veillette M, Coutu M, Richard J, Batraville LA, Dagher O, Bernard N, Tremblay C, Kaufmann DE, Roger M, Finzi A. 2015. The HIV-1 gp120 CD4-bound conformation is preferentially targeted by antibody-dependent cellular cytotoxicity-mediating antibodies in sera from HIV-1-infected individuals. J Virol 89:545–551. doi: 10.1128/JVI.02868-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alsahafi N, Ding S, Richard J, Markle T, Brassard N, Walker B, Lewis GK, Kaufmann DE, Brockman MA, Finzi A. 2015. Nef proteins from HIV-1 elite controllers are inefficient at preventing antibody-dependent cellular cytotoxicity. J Virol 90:2993–3002. doi: 10.1128/JVI.02973-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding S, Veillette M, Coutu M, Prevost J, Scharf L, Bjorkman PJ, Ferrari G, Robinson JE, Sturzel C, Hahn BH, Sauter D, Kirchhoff F, Lewis GK, Pazgier M, Finzi A. 2016. A highly conserved residue of the HIV-1 gp120 inner domain is important for antibody-dependent cellular cytotoxicity responses mediated by anti-cluster A antibodies. J Virol 90:2127–2134. doi: 10.1128/JVI.02779-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alsahafi N, Bakouche N, Kazemi M, Richard J, Ding S, Bhattacharyya S, Das D, Anand SP, Prevost J, Tolbert WD, Lu H, Medjahed H, Gendron-Lepage G, Ortega Delgado GG, Kirk S, Melillo B, Mothes W, Sodroski J, Smith AB III, Kaufmann DE, Wu X, Pazgier M, Rouiller I, Finzi A, Munro JB. 2019. An asymmetric opening of HIV-1 envelope mediates antibody-dependent cellular cytotoxicity. Cell Host Microbe 25:578–587.e5. doi: 10.1016/j.chom.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prevost J, Richard J, Medjahed H, Alexander A, Jones J, Kappes JC, Ochsenbauer C, Finzi A. 2018. Incomplete downregulation of CD4 expression affects HIV-1 Env conformation and antibody-dependent cellular cytotoxicity responses. J Virol 92:e00484-18. doi: 10.1128/JVI.00484-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arias JF, Heyer LN, von Bredow B, Weisgrau KL, Moldt B, Burton DR, Rakasz EG, Evans DT. 2014. Tetherin antagonism by Vpu protects HIV-infected cells from antibody-dependent cell-mediated cytotoxicity. Proc Natl Acad Sci U S A 111:6425–6430. doi: 10.1073/pnas.1321507111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez RA, Hamlin RE, Monroe A, Moldt B, Hotta MT, Rodriguez Caprio G, Fierer DS, Simon V, Chen BK. 2014. HIV-1 Vpu antagonism of tetherin inhibits antibody-dependent cellular cytotoxic responses by natural killer cells. J Virol 88:6031–6046. doi: 10.1128/JVI.00449-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Bredow B, Arias JF, Heyer LN, Gardner MR, Farzan M, Rakasz EG, Evans DT. 2015. Envelope glycoprotein internalization protects human and simian immunodeficiency virus-infected cells from antibody-dependent cell-mediated cytotoxicity. J Virol 89:10648–10655. doi: 10.1128/JVI.01911-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anand SP, Grover JR, Tolbert WD, Prevost J, Richard J, Ding S, Baril S, Medjahed H, Evans DT, Pazgier M, Mothes W, Finzi A. 15 May 2019, posting date Antibody-induced internalization of HIV-1 Env proteins limits surface expression of the closed conformation of Env. J Virol doi: 10.1128/JVI.00293-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richard J, Veillette M, Brassard N, Iyer SS, Roger M, Martin L, Pazgier M, Schon A, Freire E, Routy JP, Smith AB III, Park J, Jones DM, Courter JR, Melillo BN, Kaufmann DE, Hahn BH, Permar SR, Haynes BF, Madani N, Sodroski JG, Finzi A. 2015. CD4 mimetics sensitize HIV-1-infected cells to ADCC. Proc Natl Acad Sci U S A 112:E2687–E2694. doi: 10.1073/pnas.1506755112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madani N, Princiotto AM, Zhao C, Jahanbakhshsefidi F, Mertens M, Herschhorn A, Melillo B, Smith AB III, Sodroski J. 18 January 2017, posting date Activation and inactivation of primary human immunodeficiency virus envelope glycoprotein trimers by CD4-mimetic compounds. J Virol doi: 10.1128/JVI.01880-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madani N, Princiotto AM, Easterhoff D, Bradley T, Luo K, Williams WB, Liao HX, Moody MA, Phad GE, Vazquez Bernat N, Melillo B, Santra S, Smith AB III, Karlsson Hedestam GB, Haynes B, Sodroski J. 2016. Antibodies elicited by multiple envelope glycoprotein immunogens in primates neutralize primary human immunodeficiency viruses (HIV-1) sensitized by CD4-mimetic compounds. J Virol 90:5031–5046. doi: 10.1128/JVI.03211-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madani N, Princiotto AM, Schon A, LaLonde J, Feng Y, Freire E, Park J, Courter JR, Jones DM, Robinson J, Liao HX, Moody MA, Permar S, Haynes B, Smith AB III, Wyatt R, Sodroski J. 2014. CD4-mimetic small molecules sensitize human immunodeficiency virus to vaccine-elicited antibodies. J Virol 88:6542–6555. doi: 10.1128/JVI.00540-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding S, Grenier MC, Tolbert WD, Vezina D, Sherburn R, Richard J, Prevost J, Chapleau JP, Gendron-Lepage G, Medjahed H, Abrams C, Sodroski J, Pazgier M, Smith AB III, Finzi A. 26 November 2019, posting date A new family of small-molecule CD4-mimetic compounds contacts highly conserved aspartic acid 368 of HIV-1 gp120 and mediates antibody-dependent cellular cytotoxicity. J Virol doi: 10.1128/JVI.01325-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madani N, Princiotto AM, Mach L, Ding S, Prevost J, Richard J, Hora B, Sutherland L, Zhao CA, Conn BP, Bradley T, Moody MA, Melillo B, Finzi A, Haynes BF, Smith AB III, Santra S, Sodroski J. 2018. A CD4-mimetic compound enhances vaccine efficacy against stringent immunodeficiency virus challenge. Nat Commun 9:2363. doi: 10.1038/s41467-018-04758-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Princiotto AM, Vrbanac VD, Melillo B, Park J, Tager AM, Smith AB III, Sodroski J, Madani N. 2018. A small-molecule CD4-mimetic compound protects bone marrow-liver-thymus humanized mice from HIV-1 infection. J Infect Dis 218:471–475. doi: 10.1093/infdis/jiy174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiang SH, Kwong PD, Gupta R, Rizzuto CD, Casper DJ, Wyatt R, Wang L, Hendrickson WA, Doyle ML, Sodroski J. 2002. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J Virol 76:9888–9899. doi: 10.1128/jvi.76.19.9888-9899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zoubchenok D, Veillette M, Prevost J, Sanders-Buell E, Wagh K, Korber B, Chenine AL, Finzi A. 31 January 2017, posting date Histidine 375 modulates CD4 binding in HIV-1 CRF01_AE envelope glycoproteins. J Virol doi: 10.1128/JVI.02151-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prevost J, Zoubchenok D, Richard J, Veillette M, Pacheco B, Coutu M, Brassard N, Parsons MS, Ruxrungtham K, Bunupuradah T, Tovanabutra S, Hwang KK, Moody MA, Haynes BF, Bonsignori M, Sodroski J, Kaufmann DE, Shaw GM, Chenine AL, Finzi A. 2017. Influence of the envelope gp120 Phe 43 cavity on HIV-1 sensitivity to antibody-dependent cell-mediated cytotoxicity responses. J Virol 91:e02452-16. doi: 10.1128/JVI.02452-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schader SM, Colby-Germinario SP, Quashie PK, Oliveira M, Ibanescu R-I, Moisi D, Mespléde T, Wainberg MA. 2012. HIV gp120 H375 is unique to HIV-1 subtype CRF01_AE and confers strong resistance to the entry inhibitor BMS-599793, a candidate microbicide drug. Antimicrob Agents Chemother 56:4257–4267. doi: 10.1128/AAC.00639-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Wang S, Kong R, Ding W, Lee FH, Parker Z, Kim E, Learn GH, Hahn P, Policicchio B, Brocca-Cofano E, Deleage C, Hao X, Chuang GY, Gorman J, Gardner M, Lewis MG, Hatziioannou T, Santra S, Apetrei C, Pandrea I, Alam SM, Liao HX, Shen X, Tomaras GD, Farzan M, Chertova E, Keele BF, Estes JD, Lifson JD, Doms RW, Montefiori DC, Haynes BF, Sodroski JG, Kwong PD, Hahn BH, Shaw GM. 2016. Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc Natl Acad Sci U S A 113:E3413–E3422. doi: 10.1073/pnas.1606636113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melillo B, Liang S, Park J, Schon A, Courter JR, LaLonde JM, Wendler DJ, Princiotto AM, Seaman MS, Freire E, Sodroski J, Madani N, Hendrickson WA, Smith AB III.. 2016. Small-molecule CD4-mimics: structure-based optimization of HIV-1 entry inhibition. ACS Med Chem Lett 7:330–334. doi: 10.1021/acsmedchemlett.5b00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lalonde JM, Le-Khac M, Jones DM, Courter JR, Park J, Schon A, Princiotto AM, Wu X, Mascola JR, Freire E, Sodroski J, Madani N, Hendrickson WA, Smith AB III.. 2013. Structure-based design and synthesis of an HIV-1 Entry inhibitor exploiting X-ray and thermodynamic characterization. ACS Med Chem Lett 4:338–343. doi: 10.1021/ml300407y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lalonde JM, Kwon YD, Jones DM, Sun AW, Courter JR, Soeta T, Kobayashi T, Princiotto AM, Wu X, Schon A, Freire E, Kwong PD, Mascola JR, Sodroski J, Madani N, Smith AB III.. 2012. Structure-based design, synthesis, and characterization of dual hotspot small-molecule HIV-1 entry inhibitors. J Med Chem 55:4382–4396. doi: 10.1021/jm300265j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwon YD, LaLonde JM, Yang Y, Elban MA, Sugawara A, Courter JR, Jones DM, Smith AB III, Debnath AK, Kwong PD. 2014. Crystal structures of HIV-1 gp120 envelope glycoprotein in complex with NBD analogues that target the CD4-binding site. PLoS One 9:e85940. doi: 10.1371/journal.pone.0085940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwon YD, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang Y, Zhou T, Zhu J, Vicic DA, Debnath AK, Shapiro L, Bewley CA, Mascola JR, Sodroski JG, Kwong PD. 2012. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc Natl Acad Sci U S A 109:5663–5668. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Curreli F, Kwon YD, Zhang H, Yang Y, Scacalossi D, Kwong PD, Debnath AK. 2014. Binding mode characterization of NBD series CD4-mimetic HIV-1 entry inhibitors by X-ray structure and resistance study. Antimicrob Agents Chemother 58:5478–5491. doi: 10.1128/AAC.03339-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Acharya P, Luongo TS, Louder MK, McKee K, Yang Y, Kwon YD, Mascola JR, Kessler P, Martin L, Kwong PD. 2013. Structural basis for highly effective HIV-1 neutralization by CD4-mimetic miniproteins revealed by 1.5 A cocrystal structure of gp120 and M48U1. Structure 21:1018–1029. doi: 10.1016/j.str.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Madani N, Schon A, Princiotto AM, Lalonde JM, Courter JR, Soeta T, Ng D, Wang L, Brower ET, Xiang SH, Do Kwon Y, Huang CC, Wyatt R, Kwong PD, Freire E, Smith AB III, Sodroski J. 2008. Small-molecule CD4 mimics interact with a highly conserved pocket on HIV-1 gp120. Structure 16:1689–1701. doi: 10.1016/j.str.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grupping K, Selhorst P, Michiels J, Vereecken K, Heyndrickx L, Kessler P, Vanham G, Martin L, Arien KK. 2012. MiniCD4 protein resistance mutations affect binding to the HIV-1 gp120 CD4 binding site and decrease entry efficiency. Retrovirology 9:36. doi: 10.1186/1742-4690-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richard J, Veillette M, Ding S, Zoubchenok D, Alsahafi N, Coutu M, Brassard N, Park J, Courter JR, Melillo B, Smith AB III, Shaw GM, Hahn BH, Sodroski J, Kaufmann DE, Finzi A. 2016. Small CD4 mimetics prevent HIV-1 uninfected bystander CD4+ T cell killing mediated by antibody-dependent cell-mediated cytotoxicity. EBioMedicine 3:122–134. doi: 10.1016/j.ebiom.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anand SP, Prevost J, Baril S, Richard J, Medjahed H, Chapleau JP, Tolbert WD, Kirk S, Smith AB III, Wines BD, Kent SJ, Hogarth PM, Parsons MS, Pazgier M, Finzi A. 17 January 2018, posting date Two families of Env antibodies efficiently engage Fc-gamma receptors and eliminate HIV-1-infected cells. J Virol doi: 10.1128/JVI.01823-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Richard J, Prevost J, von Bredow B, Ding S, Brassard N, Medjahed H, Coutu M, Melillo B, Bibollet-Ruche F, Hahn BH, Kaufmann DE, Smith AB III, Sodroski J, Sauter D, Kirchhoff F, Gee K, Neil SJ, Evans DT, Finzi A. 2017. BST-2 expression modulates small CD4-mimetic sensitization of HIV-1-infected cells to antibody-dependent cellular cytotoxicity. J Virol 91:e00219-17. doi: 10.1128/JVI.00219-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Richard J, Pacheco B, Gohain N, Veillette M, Ding S, Alsahafi N, Tolbert WD, Prevost J, Chapleau JP, Coutu M, Jia M, Brassard N, Park J, Courter JR, Melillo B, Martin L, Tremblay C, Hahn BH, Kaufmann DE, Wu X, Smith AB III, Sodroski J, Pazgier M, Finzi A. 2016. Co-receptor binding site antibodies enable CD4-mimetics to expose conserved anti-cluster A ADCC epitopes on HIV-1 envelope glycoproteins. EBioMedicine 12:208–218. doi: 10.1016/j.ebiom.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kijak GH, Sanders-Buell E, Chenine A-L, Eller MA, Goonetilleke N, Thomas R, Leviyang S, Harbolick EA, Bose M, Pham P, Oropeza C, Poltavee K, O'Sullivan AM, Billings E, Merbah M, Costanzo MC, Warren JA, Slike B, Li H, Peachman KK, Fischer W, Gao F, Cicala C, Arthos J, Eller LA, O'Connell RJ, Sinei S, Maganga L, Kibuuka H, Nitayaphan S, Rao M, Marovich MA, Krebs SJ, Rolland M, Korber BT, Shaw GM, Michael NL, Robb ML, Tovanabutra S, Kim JH. 2017. Rare HIV-1 transmitted/founder lineages identified by deep viral sequencing contribute to rapid shifts in dominant quasispecies during acute and early infection. PLoS Pathog 13:e1006510. doi: 10.1371/journal.ppat.1006510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Derking R, Ozorowski G, Sliepen K, Yasmeen A, Cupo A, Torres JL, Julien JP, Lee JH, van Montfort T, de Taeye SW, Connors M, Burton DR, Wilson IA, Klasse PJ, Ward AB, Moore JP, Sanders RW. 2015. Comprehensive antigenic map of a cleaved soluble HIV-1 envelope trimer. PLoS Pathog 11:e1004767. doi: 10.1371/journal.ppat.1004767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma X, Lu M, Gorman J, Terry DS, Hong X, Zhou Z, Zhao H, Altman RB, Arthos J, Blanchard SC, Kwong PD, Munro JB, Mothes W. 21 March 2018, posting date HIV-1 Env trimer opens through an asymmetric intermediate in which individual protomers adopt distinct conformations. Elife doi: 10.7554/eLife.34271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao Q, Ma L, Jiang S, Lu H, Liu S, He Y, Strick N, Neamati N, Debnath AK. 2005. Identification of N-phenyl-N’-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamides as a new class of HIV-1 entry inhibitors that prevent gp120 binding to CD4. Virology 339:213–225. doi: 10.1016/j.virol.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 62.Herschhorn A, Gu C, Moraca F, Ma X, Farrell M, Smith AB III, Pancera M, Kwong PD, Schon A, Freire E, Abrams C, Blanchard SC, Mothes W, Sodroski JG. 2017. The beta20-beta21 of gp120 is a regulatory switch for HIV-1 Env conformational transitions. Nat Commun 8:1049. doi: 10.1038/s41467-017-01119-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shrivastava IH, Wendel K, LaLonde JM. 2012. Spontaneous rearrangement of the beta20/beta21 strands in simulations of unliganded HIV-1 glycoprotein, gp120. Biochemistry 51:7783–7793. doi: 10.1021/bi300878d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tolbert WD, Sherburn RT, Van V, Pazgier M. 2019. Structural basis for epitopes in the gp120 cluster A region that invokes potent effector cell activity. Viruses 11:69. doi: 10.3390/v11010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 66.Guo D, Shi X, Arledge KC, Song D, Jiang L, Fu L, Gong X, Zhang S, Wang X, Zhang L. 2012. A single residue within the V5 region of HIV-1 envelope facilitates viral escape from the broadly neutralizing monoclonal antibody VRC01. J Biol Chem 287:43170–43179. doi: 10.1074/jbc.M112.399402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou P, Wang H, Fang M, Li Y, Wang H, Shi S, Li Z, Wu J, Han X, Shi X, Shang H, Zhou T, Zhang L. 2019. Broadly resistant HIV-1 against CD4-binding site neutralizing antibodies. PLoS Pathog 15:e1007819. doi: 10.1371/journal.ppat.1007819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Otsuka Y, Schmitt K, Quinlan BD, Gardner MR, Alfant B, Reich A, Farzan M, Choe H. 2018. Diverse pathways of escape from all well-characterized VRC01-class broadly neutralizing HIV-1 antibodies. PLoS Pathog 14:e1007238. doi: 10.1371/journal.ppat.1007238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klein F, Nogueira L, Nishimura Y, Phad G, West AP Jr, Halper-Stromberg A, Horwitz JA, Gazumyan A, Liu C, Eisenreich TR, Lehmann C, Fatkenheuer G, Williams C, Shingai M, Martin MA, Bjorkman PJ, Seaman MS, Zolla-Pazner S, Karlsson Hedestam GB, Nussenzweig MC. 2014. Enhanced HIV-1 immunotherapy by commonly arising antibodies that target virus escape variants. J Exp Med 211:2361–2372. doi: 10.1084/jem.20141050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, Zang T, Dorner M, Billerbeck E, Labitt RN, Gaebler C, Marcovecchio PM, Incesu RB, Eisenreich TR, Bieniasz PD, Seaman MS, Bjorkman PJ, Ravetch JV, Ploss A, Nussenzweig MC. 2012. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 492:118–122. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Caskey M, Klein F, Lorenzi JCC, Seaman MS, West AP, Buckley N, Kremer G, Nogueira L, Braunschweig M, Scheid JF, Horwitz JA, Shimeliovich I, Ben-Avraham S, Witmer-Pack M, Platten M, Lehmann C, Burke LA, Hawthorne T, Gorelick RJ, Walker BD, Keler T, Gulick RM, Fätkenheuer G, Schlesinger SJ, Nussenzweig MC. 2015. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522:487–491. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuan T, Li J, Zhang MY. 2013. HIV-1 envelope glycoprotein variable loops are indispensable for envelope structural integrity and virus entry. PLoS One 8:e69789. doi: 10.1371/journal.pone.0069789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rashad AA, Acharya K, Haftl A, Aneja R, Dick A, Holmes AP, Chaiken I. 2017. Chemical optimization of macrocyclic HIV-1 inactivators for improving potency and increasing the structural diversity at the triazole ring. Org Biomol Chem 15:7770–7782. doi: 10.1039/c7ob01448a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rashad AA, Kalyana Sundaram RV, Aneja R, Duffy C, Chaiken I. 2015. Macrocyclic envelope glycoprotein antagonists that irreversibly inactivate HIV-1 before host cell encounter. J Med Chem 58:7603–7608. doi: 10.1021/acs.jmedchem.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kassa A, Finzi A, Pancera M, Courter JR, Smith AB III, Sodroski J. 11 February 2009, posting date Identification of a human immunodeficiency virus (HIV-1) envelope glycoprotein variant resistant to cold inactivation. J Virol doi: 10.1128/JVI.02110-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kassa A, Madani N, Schon A, Haim H, Finzi A, Xiang SH, Wang L, Princiotto A, Pancera M, Courter J, Smith AB III, Freire E, Kwong PD, Sodroski J. 2009. Transitions to and from the CD4-bound conformation are modulated by a single-residue change in the human immunodeficiency virus type 1 gp120 inner domain. J Virol 83:8364–8378. doi: 10.1128/JVI.00594-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.DeLeon O, Hodis H, O'Malley Y, Johnson J, Salimi H, Zhai Y, Winter E, Remec C, Eichelberger N, Van Cleave B, Puliadi R, Harrington RD, Stapleton JT, Haim H. 2017. Accurate predictions of population-level changes in sequence and structural properties of HIV-1 Env using a volatility-controlled diffusion model. PLoS Biol 15:e2001549. doi: 10.1371/journal.pbio.2001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fontaine J, Chagnon-Choquet J, Valcke HS, Poudrier J, Roger M; Montreal Primary HIV Infection and Long-Term Non-Progressor Study Groups. 2011. High expression levels of B lymphocyte stimulator (BLyS) by dendritic cells correlate with HIV-related B-cell disease progression in humans. Blood 117:145–155. doi: 10.1182/blood-2010-08-301887. [DOI] [PubMed] [Google Scholar]
- 79.Fontaine J, Coutlee F, Tremblay C, Routy JP, Poudrier J, Roger M; Montreal Primary HIV Infection and Long-Term Nonprogressor Study Groups. 2009. HIV infection affects blood myeloid dendritic cells after successful therapy and despite nonprogressing clinical disease. J Infect Dis 199:1007–1018. doi: 10.1086/597278. [DOI] [PubMed] [Google Scholar]
- 80.Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PIW, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, Kadie CM, Carlson JM, Heckerman D, Graham RR, Plenge RM, Deeks SG, Gianniny L, Crawford G, Sullivan J, Gonzalez E, Davies L, Camargo A, Moore JM, Beattie N, Gupta S, Crenshaw A, Burtt NP, Guiducci C, Gupta N, Gao X, Qi Y, Yuki Y, Piechocka-Trocha A, Cutrell E, Rosenberg R, Moss KL, Lemay P, O'Leary J, Schaefer T, Verma P, Toth I, Block B, Baker B, Rothchild A, Lian J, Proudfoot J, Alvino DML, Vine S, Addo MM, Allen TM, et al. . 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kamya P, Boulet S, Tsoukas CM, Routy JP, Thomas R, Cote P, Boulassel MR, Baril JG, Kovacs C, Migueles SA, Connors M, Suscovich TJ, Brander C, Tremblay CL, Bernard N; Canadian Cohort of HIV Infected Slow Progressors. 2011. Receptor-ligand requirements for increased NK cell polyfunctional potential in slow progressors infected with HIV-1 coexpressing KIR3DL1*h/*y and HLA-B*57. J Virol 85:5949–5960. doi: 10.1128/JVI.02652-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peretz Y, Ndongala ML, Boulet S, Boulassel MR, Rouleau D, Cote P, Longpre D, Routy JP, Falutz J, Tremblay C, Tsoukas CM, Sekaly RP, Bernard NF. 2007. Functional T cell subsets contribute differentially to HIV peptide-specific responses within infected individuals: correlation of these functional T cell subsets with markers of disease progression. Clin Immunol 124:57–68. doi: 10.1016/j.clim.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 83.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Korber B, Foley BT, Kuiken C, Pillai SK, Sodroski JG. 1998. Numbering positions in HIV relative to HXB2CG, p 102–111. In Korber B, Kuiken CL, Foley B, Hahn B, McCutchan F, Mellors JW, Sodroski J (ed), Human Retroviruses and AIDS 1998. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM. [Google Scholar]
- 85.Eddy SR. 20 October 2011, posting date Accelerated profile HMM searches. PLoS Comput Biol doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 87.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard NP, Gerard C, Sodroski J, Choe H. 1999. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 88.Montefiori DC, Karnasuta C, Huang Y, Ahmed H, Gilbert P, de Souza MS, McLinden R, Tovanabutra S, Laurence-Chenine A, Sanders-Buell E, Moody MA, Bonsignori M, Ochsenbauer C, Kappes J, Tang H, Greene K, Gao H, LaBranche CC, Andrews C, Polonis VR, Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Self SG, Berman PW, Francis D, Sinangil F, Lee C, Tartaglia J, Robb ML, Haynes BF, Michael NL, Kim JH. 2012. Magnitude and breadth of the neutralizing antibody response in the RV144 and Vax003 HIV-1 vaccine efficacy trials. J Infect Dis 206:431–441. doi: 10.1093/infdis/jis367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu X, Parast AB, Richardson BA, Nduati R, John-Stewart G, Mbori-Ngacha D, Rainwater SM, Overbaugh J. 2006. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J Virol 80:835–844. doi: 10.1128/JVI.80.2.835-844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lodge R, Lalonde JP, Lemay G, Cohen EA. 1997. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J 16:695–705. doi: 10.1093/emboj/16.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chenine AL, Merbah M, Wieczorek L, Molnar S, Mann B, Lee J, O’Sullivan AM, Bose M, Sanders-Buell E, Kijak GH, Herrera C, McLinden R, O’Connell RJ, Michael NL, Robb ML, Kim JH, Polonis VR, Tovanabutra S. 2018. Neutralization sensitivity of a novel HIV-1 CRF01_AE panel of infectious molecular clones. J Acquir Immune Defic Syndr 78:348–355. doi: 10.1097/QAI.0000000000001675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, Parrish NF, Shaw KS, Guffey MB, Bar KJ, Davis KL, Ochsenbauer-Jambor C, Kappes JC, Saag MS, Cohen MS, Mulenga J, Derdeyn CA, Allen S, Hunter E, Markowitz M, Hraber P, Perelson AS, Bhattacharya T, Haynes BF, Korber BT, Hahn BH, Shaw GM. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med 206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ochsenbauer C, Edmonds TG, Ding H, Keele BF, Decker J, Salazar MG, Salazar-Gonzalez JF, Shattock R, Haynes BF, Shaw GM, Hahn BH, Kappes JC. 2012. Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J Virol 86:2715–2728. doi: 10.1128/JVI.06157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, Haynes BF; NISC Comparative Sequencing Program. 2013. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bar KJ, Tsao CY, Iyer SS, Decker JM, Yang Y, Bonsignori M, Chen X, Hwang KK, Montefiori DC, Liao HX, Hraber P, Fischer W, Li H, Wang S, Sterrett S, Keele BF, Ganusov VV, Perelson AS, Korber BT, Georgiev I, McLellan JS, Pavlicek JW, Gao F, Haynes BF, Hahn BH, Kwong PD, Shaw GM. 2012. Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape. PLoS Pathog 8:e1002721. doi: 10.1371/journal.ppat.1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fenton-May AE, Dibben O, Emmerich T, Ding H, Pfafferott K, Aasa-Chapman MM, Pellegrino P, Williams I, Cohen MS, Gao F, Shaw GM, Hahn BH, Ochsenbauer C, Kappes JC, Borrow P. 2013. Relative resistance of HIV-1 founder viruses to control by interferon-alpha. Retrovirology 10:146. doi: 10.1186/1742-4690-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, Hora B, Berg A, Cai F, Hopper J, Denny TN, Ding H, Ochsenbauer C, Kappes JC, Galimidi RP, West AP, Bjorkman PJ, Wilen CB, Doms RW, O'Brien M, Bhardwaj N, Borrow P, Haynes BF, Muldoon M, Theiler JP, Korber B, Shaw GM, Hahn BH. 2013. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci U S A 110:6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gao F, Bonsignori M, Liao HX, Kumar A, Xia SM, Lu X, Cai F, Hwang KK, Song H, Zhou T, Lynch RM, Alam SM, Moody MA, Ferrari G, Berrong M, Kelsoe G, Shaw GM, Hahn BH, Montefiori DC, Kamanga G, Cohen MS, Hraber P, Kwong PD, Korber BT, Mascola JR, Kepler TB, Haynes BF. 2014. Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell 158:481–491. doi: 10.1016/j.cell.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.O'Brien WA, Koyanagi Y, Namazie A, Zhao JQ, Diagne A, Idler K, Zack JA, Chen IS. 1990. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature 348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 100.Rho HM, Poiesz B, Ruscetti FW, Gallo RC. 1981. Characterization of the reverse transcriptase from a new retrovirus (HTLV) produced by a human cutaneous T-cell lymphoma cell line. Virology 112:355–360. doi: 10.1016/0042-6822(81)90642-5. [DOI] [PubMed] [Google Scholar]
- 101.Martin L, Stricher F, Misse D, Sironi F, Pugniere M, Barthe P, Prado-Gotor R, Freulon I, Magne X, Roumestand C, Menez A, Lusso P, Veas F, Vita C. 2003. Rational design of a CD4 mimic that inhibits HIV-1 entry and exposes cryptic neutralization epitopes. Nat Biotechnol 21:71–76. doi: 10.1038/nbt768. [DOI] [PubMed] [Google Scholar]
- 102.Schon A, Madani N, Klein JC, Hubicki A, Ng D, Yang X, Smith AB III, Sodroski J, Freire E. 2006. Thermodynamics of binding of a low-molecular-weight CD4 mimetic to HIV-1 gp120. Biochemistry 45:10973–10980. doi: 10.1021/bi061193r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Prevost J, Richard J, Ding S, Pacheco B, Charlebois R, Hahn BH, Kaufmann DE, Finzi A. 2018. Envelope glycoproteins sampling states 2/3 are susceptible to ADCC by sera from HIV-1-infected individuals. Virology 515:38–45. doi: 10.1016/j.virol.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 105.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 106.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 107.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Acharya P, Tolbert WD, Gohain N, Wu X, Yu L, Liu T, Huang W, Huang CC, Kwon YD, Louder RK, Luongo TS, McLellan JS, Pancera M, Yang Y, Zhang B, Flinko R, Foulke JS Jr, Sajadi MM, Kamin-Lewis R, Robinson JE, Martin L, Kwong PD, Guan Y, DeVico AL, Lewis GK, Pazgier M. 2014. Structural definition of an antibody-dependent cellular cytotoxicity response implicated in reduced risk for HIV-1 infection. J Virol 88:12895–12906. doi: 10.1128/JVI.02194-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect of single gp120 layer mutations on neutralization by CD4 and CD4mc. Recombinant HIV-1 strains expressing luciferase and bearing wild-type or mutant CRF01_AE Envs (92TH023 and CM244 isolates) were normalized by reverse transcriptase activity. Normalized amounts of viruses were incubated with serial dilutions of sCD4 (A and B), BNM-III-170 (C and D), or M48U1 (E and F) at 37°C for 1 h prior to infection of Cf2Th-CD4/CCR5 cells. Infectivity at each dilution of sCD4 or CD4mc tested is shown as the percentage of infection without sCD4 or CD4mc for each particular mutant. Quadruplicate samples were analyzed in each experiment. Data shown are the means of results obtained in at least 3 independent experiments. The error bars represent the standard deviations. Neutralization half-maximal inhibitory concentrations (IC50) are summarized in panel G. Download FIG S1, TIF file, 0.5 MB (481.8KB, tif) .
Copyright © 2020 Prévost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phe43 cavity and inner domain changes render the CRF01_AE HIV-1 strain susceptible to CD4mc-induced Env conformational changes. Cell surface staining of 293T cells transfected with different Env expressors (92TH023, CM244, JRFL, YU2, and BG505 isolates [WT or their mutated counterparts]) was performed using a panel of Env ligands. Binding of bNAbs (panels A and B and panels E to G) and nnAbs (panels C and D and panels H to J) was performed in the presence of BNM-III-170 (50 μM) or in its absence (DMSO). Shown are the mean fluorescence intensities (MFI) obtained in the presence of BNM-III-170 normalized to the MFI or in absence of BNM-III-170 (DMSO) from the transfected (GFP+) population for staining obtained in at least 3 independent experiments. All MFI data were normalized to 2G12 MFI for each Env mutant. Error bars indicate means ± SEM. Statistical significance was tested using an unpaired t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant). Download FIG S2, TIF file, 0.5 MB (473.4KB, tif) .
Copyright © 2020 Prévost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Electron density maps of CD4mc in complex with gp120. A 2Fo-Fc electron density map contoured at 1 s shows the density around MCG-IV-210 for the LM+HS gp12093TH057 coree (left) and the LM+HT gp12093TH057 coree (right). MCG-IV-210 is colored gray and gp120 green. Nitrogens are colored blue and oxygens red. Download FIG S3, TIF file, 2.9 MB (2.9MB, tif) .
Copyright © 2020 Prévost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
X-ray crystallography data collection and refinement statistics. Download Table S1, DOCX file, 0.02 MB (17.4KB, docx) .
Copyright © 2020 Prévost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phe43 cavity and inner domain changes enhance the sensitivity of CRF01_AE strains to neutralization by cPTs. (A) Chemical structure of the different cPTs tested. (B to D) Recombinant HIV-1 strains expressing luciferase and bearing wild-type or mutant CRF01_AE Envs (92TH023 isolate) were normalized by reverse transcriptase activity. Normalized amounts of viruses were incubated with serial dilutions of the cyclic peptide triazoles (cPTs) AAR029N2 (B) and AAR029N3 (C) at 37°C for 1 h prior to infection of Cf2Th-CD4/CCR5 cells. Infectivity at each dilution of cPTs tested is shown as the percentage of infection without cPTs for each particular mutant. Quadruplicate samples were analyzed in each experiment. Data shown are the means of results obtained in at least 3 independent experiments. The error bars represent the standard deviations. Neutralization half-maximal inhibitory concentrations (IC50) are summarized in panel D. Download FIG S4, TIF file, 0.3 MB (282.4KB, tif) .
Copyright © 2020 Prévost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Details about chemical synthesis. Download Text S1, DOCX file, 0.8 MB (809.9KB, docx) .
Copyright © 2020 Prévost et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.