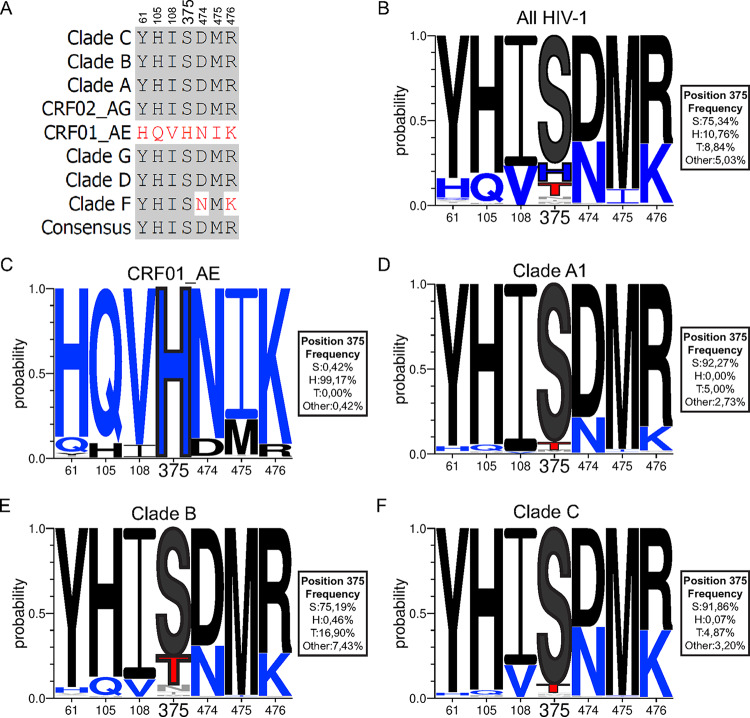
FIG 1.
Sequence alignment of gp120 Phe43 cavity and inner domain layer residues of different HIV-1 isolates. (A) Sequence alignment of selected gp120 residues located in the Phe43 cavity (375) or the inner domain layers (61, 105, 108, 474, 475, and 476) based on the Env consensus sequence of each of the HIV-1 group M major subtypes (clades A, B, C, D, F, G, CRF01_AE, and CRF02_AG). The 2017 Los Alamos database-curated Env alignment was used as the basis for this figure, which contains 5,471 amino acid HIV-1 group M sequences (including 481 sequences of CRF01_AE, 220 of subtype A1, 1,937 of subtype B, and 1,377 of subtype C). Residue numbering is based on that of the HXBc2 strain of HIV-1. Identical residues are shaded in dark gray, and nonidentical residues are highlighted in red. (B to F) Logo depictions of the frequency of each amino acid from the Phe43 cavity at positions 61, 105, 108, 375, 474, 475, and 476 in all HIV-1 isolates (B) or in isolates from CRF01_AE (C), clade A1 (D), clade B (E), and clade C (F). The height of the letter indicates its frequency within the clade. S375 and its coevolving residues are shown in black, H375 and its coevolving residues are shown in blue, and T375 is shown in red. The box beside each logo indicates the frequency of all the amino acids at position 375.