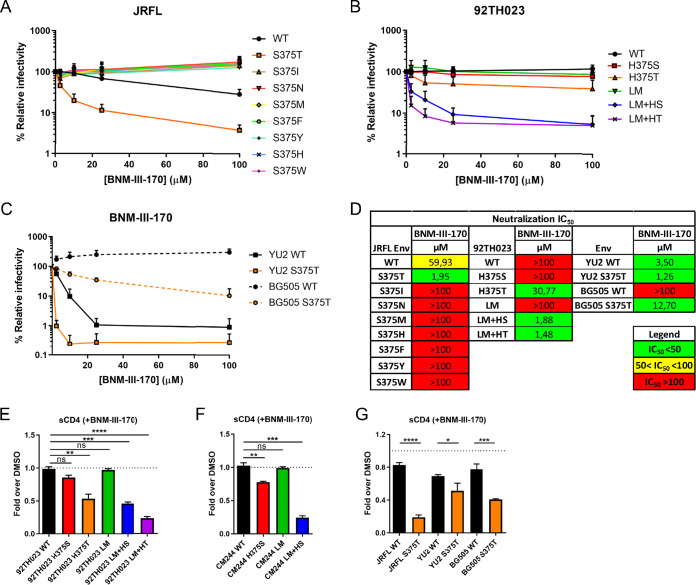
FIG 3.
Effect of Env residue 375 on CD4mc neutralization sensitivity. (A to D) Recombinant HIV-1 strains expressing luciferase and bearing wild-type or mutant Envs (JRFL, 92TH023, YU2, and BG505 isolates) were normalized by reverse transcriptase activity. Normalized amounts of viruses were incubated with serial dilutions of BNM-III-170 (A to C) at 37°C for 1 h prior to infection of Cf2Th-CD4/CCR5 cells. Infectivity at each dilution of CD4mc tested is shown as the percentage of infection without CD4mc for each particular mutant. Quadruplicate samples were analyzed in each experiment. Data shown are the means of results obtained in at least 3 independent experiments. The error bars represent the standard deviations. Neutralization half-maximal inhibitory concentrations (IC50) are summarized in panel D. (E to G) Cell surface staining of 293T cells transfected with different Env expressors (92TH023, CM244, JRFL, YU2, and BG505 isolates [WT or their mutated counterparts]). Binding of sCD4 in the presence of BNM-III-170 (50 μM) or in its absence (DMSO) was detected with the anti-CD4 OKT4 MAb. Shown are the mean fluorescence intensities (MFI) obtained in the presence of BNM-III-170 normalized to the MFI in the absence of BNM-III-170 (DMSO) from the transfected (GFP+) population for staining obtained in at least 3 independent experiments. All MFI data were normalized to 2G12 MFI for each Env mutant. Error bars indicate means ± standard errors of the means (SEM). Statistical significance was tested using an unpaired t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant).