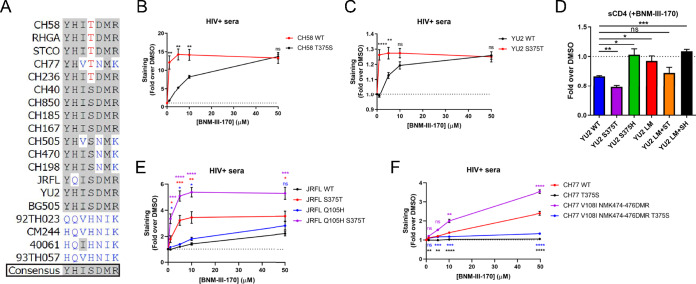
FIG 6.
Phe43 cavity and inner domain residues modulate the susceptibility of clade B strains to CD4mc. (A) Sequence alignment of selected gp120 residues located in the Phe43 cavity (375) or the inner domain layers (61, 105, 108, 474, 475, and 476) for all HIV-1 strains used in this study. S375 and its coevolving residues are shown in black, H375 and its coevolving residues are shown in blue, and T375 is shown in red. Residues corresponding to the HIV-1 consensus sequence are shaded in gray. (B, C, E, and F) Cell surface staining with HIV+ sera of primary CD4+ T cells infected with clade B primary viruses (CH58, YU2, JRFL, and CH77) or their variants in the presence of increasing concentrations of BNM-III-170 (0, 1, 5, 10, and 50 μM). The graphs shown represent the compiled mean fluorescence intensities (MFI) obtained with 5 different HIV+ sera in the presence of BNM-III-170 normalized to the MFI in the absence of BNM-III-170 (DMSO) from the infected (p24+) population. These results were obtained in at least 2 independent experiments. (D) Cell surface staining of 293T cells transfected with an expressor of the HIV-1YU2 Env WT or its mutated counterparts. Binding of sCD4 in the presence of BNM-III-170 (50 μM) or in its absence (DMSO) was detected with the anti-CD4 OKT4 MAb. Shown are the mean fluorescence intensities (MFI) obtained in the presence of BNM-III-170 normalized to the MFI in the absence of BNM-III-170 (DMSO) from the transfected (GFP+) population for staining obtained in at least 3 independent experiments. All MFI data were normalized to 2G12 MFI for each Env mutant. Error bars indicate means ± SEM. Statistical significance was tested using an unpaired t test or Mann-Whitney U test based on statistical normality (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant).