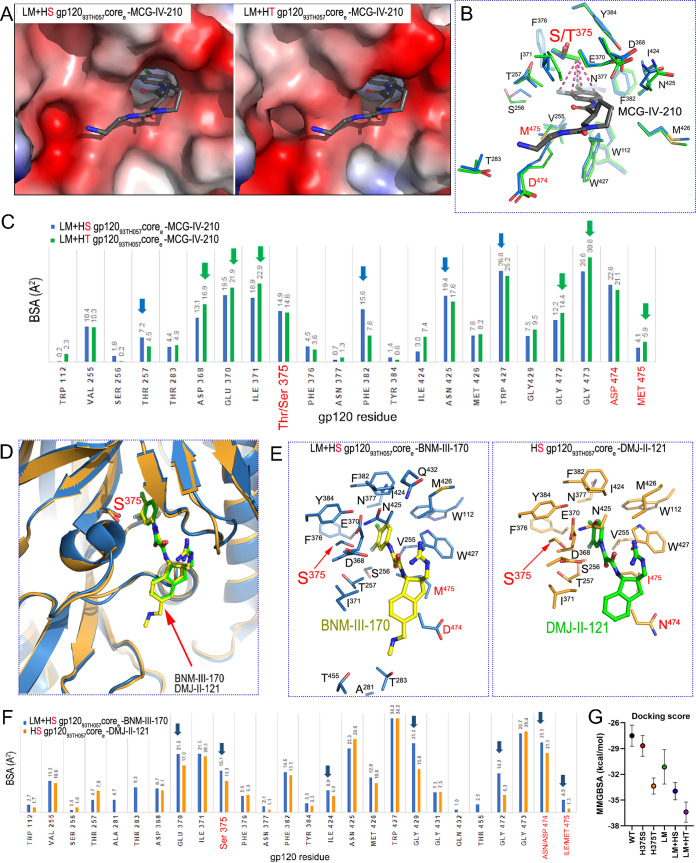
FIG 8.
Structural effects of Phe43 cavity and inner domain changes on CD4mc docking into the Phe43 cavity. (A) Magnified views of the CD4 binding pocket of LM+HS and LM+HT gp12093TH057 core with MCG-IV-210 bound. Complexes are superimposed based on the gp120 core, and electrostatic surfaces are displayed over the gp120 molecule, with blue for electropositive and red for electronegative. The MGC-IV-210 compound is shown as balls and sticks. The LM+HT gp12093TH057 core–MCG-IV-210 complex shown is from PDB ID 6P9N. (B) Details of MCG-IV-210 interactions within the binding pocket. The side chains of gp120 residues that interact with MCG-IV-210 are shown as sticks; LM+HS is depicted in green and LM+HT in blue. MCG-IV-210 is shown with balls and sticks. MCG-IV-210 atoms that interact with the T375 γ-carbon methyl group are shown with dashes connecting to the methyl group. (C) Analysis of the MCG-IV-210 interface for binding to LM+HS and LM+HT gp120. The relative contributions of gp120 residues to compound binding are shown as buried surface area (BSA) data as calculated by PISA. BSA data represent the solvent-accessible surface area of the corresponding residue that is buried upon interface formation. (D) Structural comparison of LM+HS gp12093TH057 coree–BNM-III-170 and HS gp12093TH057 coree–DMJ-II-121 (PDB ID 4I54) complex structures. Complexes are superimposed based on the gp120 core. CD4mc compounds and Ser375 are shown as sticks. (E) Details of binding of BNM-III-170 (left panel) and DMJ-II-121 (right panel) to LM+HS gp12093TH057 coree and HS gp12093TH057 coree, respectively. Residues forming the binding site for the CD4mc compound are shown as sticks (with Gly residues omitted) with LM residues labeled in red. (F) Analysis of the gp120 corees-CD4mc binding interfaces. The relative contributions of gp120 residues to BNM-III-170 and DMJ-II-121 binding are shown by the buried surface area (BSA) as calculated by PISA. BSA represents the solvent-accessible surface area of the corresponding residue that is buried upon interface formation. (G) Docking score based on MMGBSA interaction energies, showing five replica simulations each along with averages, for six BNM-III-170-bound gp120 models.