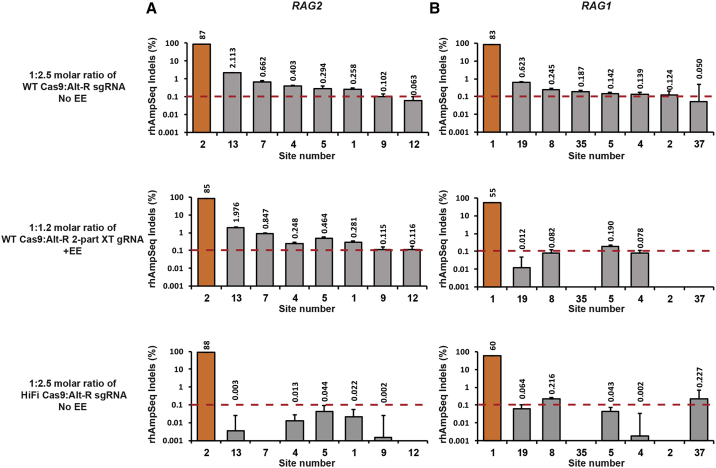
Figure 4.
rhAmpSeq-Quantified Editing in RAG2 and RAG1 Off-Target Sites in CD34+ HSPCs
(A and B) RAG2 (A) and RAG1 (B) on- and off-target editing levels were determined by rhAmpSeq in CD34+ HSPCs. WT Cas9 was complexed with either Alt-R sgRNA at a 1:2.5 Cas9:gRNA molar ratio (top plots) or with Alt-R 2-part XT gRNA at a 1:1.2 Cas9:gRNA (middle plots). Only 2-part XT RNPs were delivered in the presence of Alt-R EE. Alt-R HiFi Cas9 was complexed with sgRNA at a 1:2.5 Cas9:gRNA molar ratio (bottom plots). rhAmpSeq data were analyzed using a custom-built pipeline (IDT). Sites with editing levels higher than 0.1% in at least one of the experimental set-ups are presented (0.1% threshold designated by red dashed lines). Editing percentages are designated above the bars, and the numbers of the identified off-target sites appear below the graphs. On-target bars are colored in orange, and off-target bars are colored in gray. Error bars represent a 95% confidence interval (see Materials and Methods).