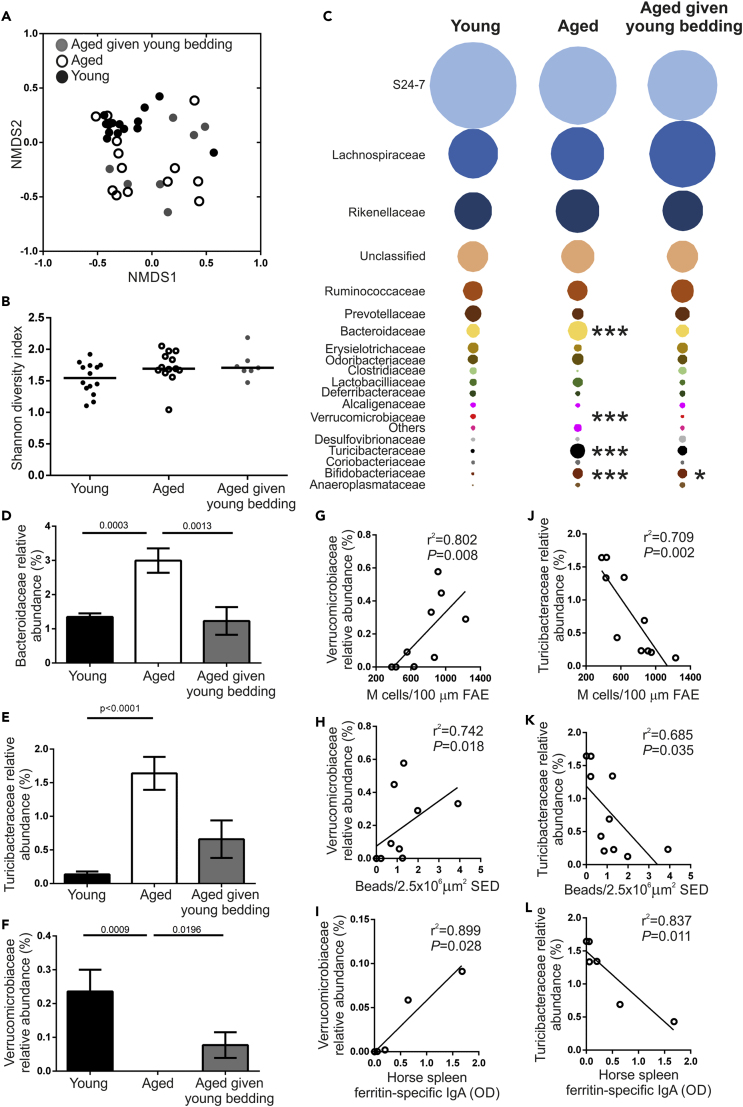
Figure 3.
Changes to the Microbiota after Housing Aged Mice on Used Bedding from Young Mice
(A) NMDS plot comparing fecal microbiota structure between mice from each age group. n = 7–14/group. Fecal microbiotas were significantly different between young mice and each group of aged mice (P = 0.002; HOMOVA).
(B) Bacterial diversity in feces from young, aged, and aged mice given young bedding was compared using the Shannon index. Horizontal line, median. n = 7–14/group. Statistical differences determined by Kruskal-Wallis.
(C) Bubble plot showing the relative abundance of distinct bacterial families in the fecal microbiotas of mice from each group. n = 7–14/group. Statistical differences for each family determined by one-way ANOVA or Kruskal-Wallis where appropriate. ∗ P < 0.05; ∗∗∗ P < 0.001.
(D–F) The relative abundance of (D) Bifidobacteriaceae, (E) Turicibacteriaceae, and (F) Verrucomicrobiaceae in feces of young, aged, and aged mice given young bedding. Data presented as mean ± SEM. n = 7–14/group. Statistical differences determined by Kruskal-Wallis.
(G–I) Correlation between the relative abundance of Verrucomicrobiaceae in feces and (G) the density of FAE GP2+ M cells, (H) the number of fluorescent 200-nm nanobeads in the SED, and (I) the level of antigen-specific fecal IgA. Statistical significance determined by Spearman correlation.
(J–L) Correlation between the relative abundance of Turicibacteriaceae in the feces and (J) the density of FAE GP2+ M cells, (K) the number of fluorescent 200-nm nanobeads detected in the SED, and (L) the level of antigen-specific fecal IgA. Statistical significance determined by Spearman correlation.