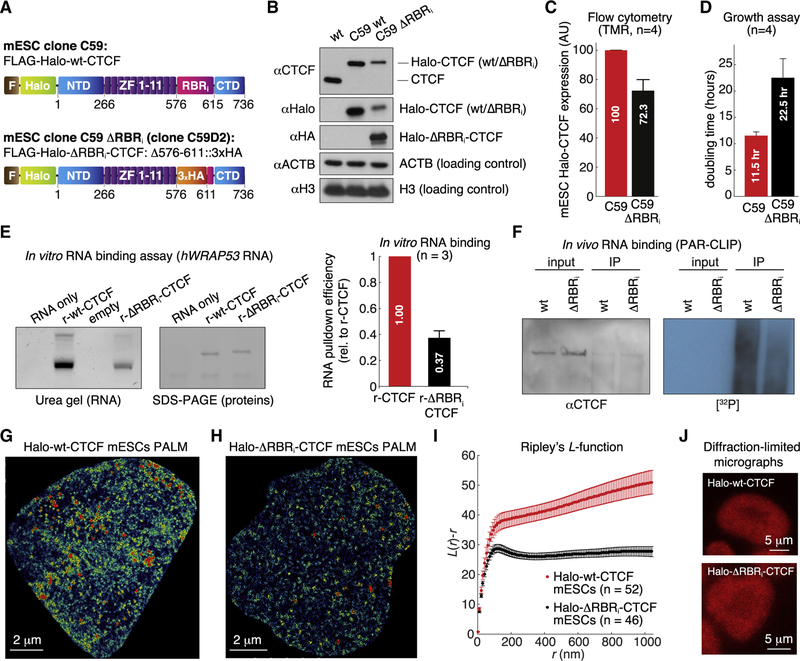
Figure 2. CTCF RBRi Region Mediates CTCF Clustering.
(A) CTCF domains in the mESC clones C59 (Halo-WT CTCF) and C59 ΔRBRi (Halo-ΔRBRi CTCF).
(B) Western blot of total cell lysates from JM8.N4 WT mESCs, C59, and C59 ΔRBRi. WT-CTCF and ΔRBRi-CTCF have the same number of amino acids, but ΔRBRi-CTCF runs slightly slower in BisTris SDS-PAGE.
(C) Flow cytometry measurement of Halo-CTCF abundance in live C59 Halo-WT CTCF and C59 ΔRBRi mESCs after TMR labeling.
(D) Growth assay for C59 Halo-WT CTCF and C59 ΔRBRi mESCs. In (C) and (D), error bars indicate mean and SE (n = 4).
(E) In vitro RNA-binding assay. An in vitro-transcribed fragment of human WRAP53 mRNA (hWRAP53, nucleotides 1–167) was incubated with recombinant (r-) WT- or ΔRBRi-CTCF protein (see STAR Methods). Recovered RNA was run on urea denaturing gels and stained with SYBR Gold; recovered proteins were run on SDS-PAGE and stained with PageBlue. Left: representative experiment (replicates in Figure S1J). Right: RNA binding efficiency of WT- versus ΔRBRi-CTCF averaged across three experiments, normalized by recovered proteins.
(F) PAR-CLIP of WT-CTCF and ΔRBRi-CTCF mESCs. Left: western blot of input and CTCF-IP. Right: autoradiography for 32P-labeled RNA for input and CTCF-IP.
(G and H) Representative PALM reconstructions for Halo-WT CTCF (G) and Halo-ΔRBRi CTCF (H).
(I) Ripley’s L function for WT-CTCF (52 cells) and ΔRBRi-CTCF mESCs (46 cells) (mean and SE).
(J) Representative confocal micrographs of mESC colonies. Halo-WT CTCF and Halo-ΔRBRi CTCF mESCs were labeled with 500 nM Halo-TMR dye and visualized using a Zeiss LSM 710 laser scanning confocal microscope.
See also Figure S1.