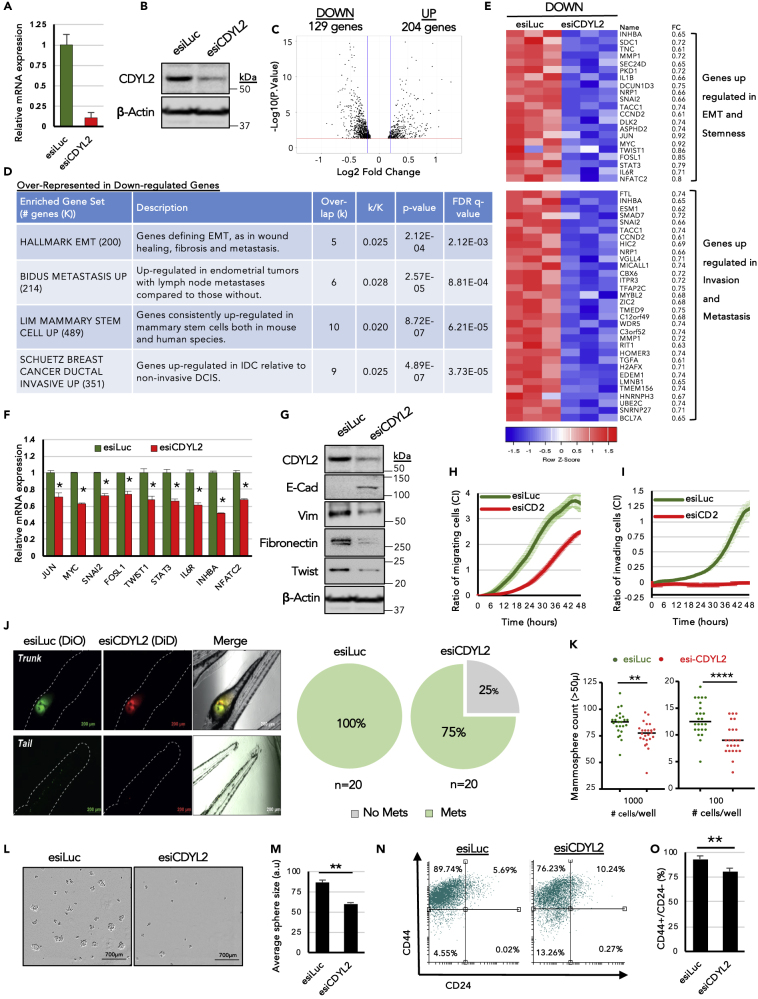
Figure 4.
RNAi Knockdown of CDYL2 in the Invasive Breast Cancer Cell Line MDA-MB-231 Induces Transcriptional and Phenotypic Changes Associated with Inhibition of Malignancy
(A and B) CDYL2 knock-down validated by RT-qPCR (A) and western blotting (B).
(C and D) (C) Volcano plot showing genes up- or down-regulated at least 1.25-fold. (D) Selected molecular signatures from the MSigDB database that were over-represented in either the up- or down-regulated gene sets from (C).
(E) Heatmap showing expression of selected genes from (D) in the triplicate RNA-seq.
(F) qRT-PCR validation of differential expression of selected genes from (D). Expression normalized to GAPDH. Data are the mean ± SD of three independent experiments. All differential expressions significant at p < 0.05 (t test).
(G) Western blot analysis of a panel of EMT markers, CDYL2, and beta-actin.
(H) xCELLigence assay comparing the relative migration efficiency of MDA-MB-231 cells treated with esiLuc or esiCDYL2 (esiCD2). Both (H) and (I) show technical quadruplicates ± SD. Experiments were repeated at least three times with similar results.
(I) Invasion assays were performed as in (H), except that the porous membrane separating the upper and lower chambers of the transwell was first overlaid with Matrigel.
(J) Fluorescence microscopy analysis of with esiLuc or esiCDYL2 on the migration of MDA-MB-231 from the perivitelline space of zebrafish embryos to the tail. Representative images are shown. Scale bar, 700 μM. Quantification of the percentage of embryos exhibiting tail metastases (Mets) is shown to the right. Experiments were repeated three times with similar results.
(K) Mammosphere formation in MDA-MB-231 cells treated with esiLuc or esiCDYL2. Cells were plated at the indicated seeding number per well in 96-well plates. Mammospheres with size >50 μm were counted after 8 days. Shown is a scatterplot of the results of a representative of three independent experiments indicating the median (black bar) and t test significance (∗∗p < 0.01; ∗∗∗∗p < 0.0001).
(L) Representative 4× phase microscopy images of mammospheres counted in (K) are shown. Scale bar, 200 μM.
(M) The diameters of mammospheres from (K) were determined using image analysis software. Shown is the mean +SD of eight wells in which 1,000 cells were seeded. t test significance (∗∗p < 0.01).
(N and O) FACS analysis of antigenic profile associated with breast cancer stem cells (CD44+; CD24-low/negative). Shown are representative FACS scatterplots (N) and the mean of three independent experiments ± SD. (O). t test significance (∗∗p < 0.01).