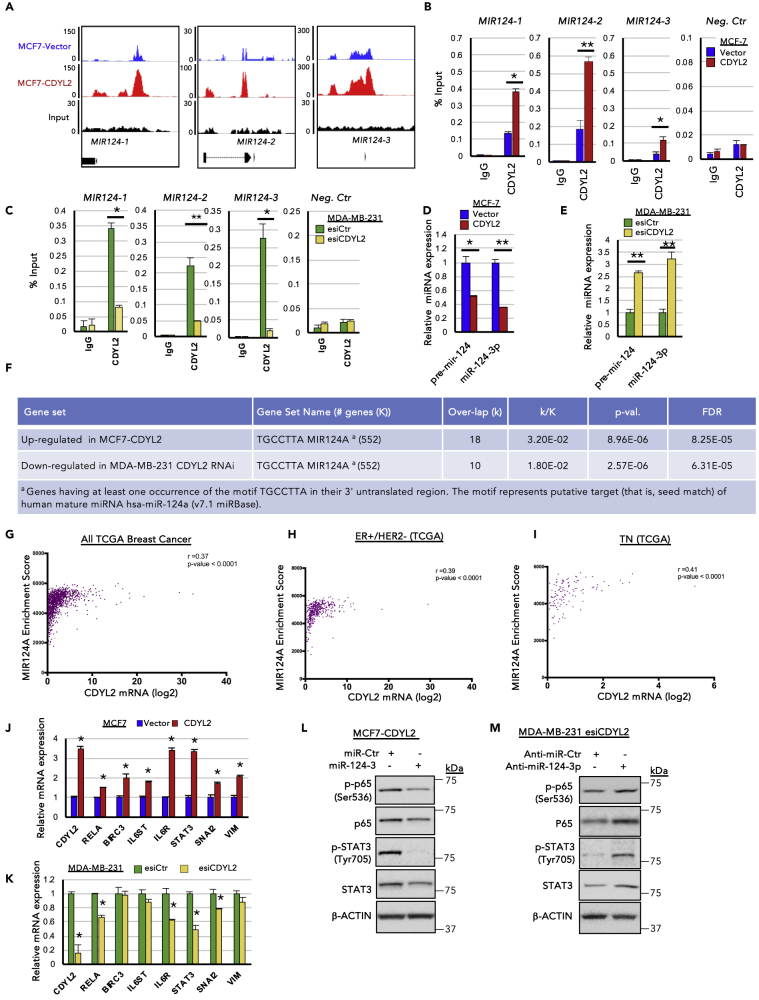
Figure 6.
CDYL2 ChIP-Seq Analysis Identifies miR-124 as a Mediator of CDYL2 Regulation of STAT3 and NF-κB Signaling
(A) Relative enrichment of CDYL2 upstream of MIR124 genes in MCF7-Vector cells (blue) and MCF7-CDYL2 (red), as revealed by ChIP-seq analysis. Input control and gene positions relative to the peaks are shown below.
(B) ChIP-qPCR validation of data presented in (A). ChIP-qPCR signal at an unrelated negative control sequence is also shown. Shown is the mean enrichment as a percentage of input of three independent experiments, ± SD. (∗p < 0.05; ∗∗p < 0.01, t test)
(C) As in (B), except CDYL2 or IgG ChIP was performed using chromatin prepared from MDA-MB-231 cells treated with esiLuc or esiCDYL2.
(D) qRT-PCR analysis of pre-mir-124 and miR-124-3p levels in MCF7-CDYL2 and MCF7-Vector cells. Expression was normalized to an unrelated miRNA. Data represent the mean of three independent experiments ± SD. Significance determined by test (∗p < 0.05; ∗∗p < 0.01).
(E) As in (D), except qRT-PCR analysis was performed using microRNA (miRNA) prepared from MDA- MB-231 cells treated with esiLuc or esiCDYL2.
(F) Selected gene expression signatures enriched in the indicated RNA-seq datasets.
(G–I) Correlation between CDYL2 expression and the GSEA signature “TGCCTTA MIR124A” in the indicated TCGA breast cancer patient cohorts. The linear regression r and p value are indicated.
(J) qRT-PCR analysis of the expression of miR-124-3p target genes in MCF7-CDYL2 and MCF7-Vector cells. Data represented as mean of three independent experiments ± SD. (∗p < 0.05, by t test).
(K) As in (J), except qRT-PCR analysis was performed using RNA prepared from MDA-MB-231 cells treated with esiLuc or esiCDYL2.
(L and M) Western blot of phosphorylated p65 (Ser 536), total p65, phosphorylated STAT3 (Tyr 705), and total STAT3 in MCF7-CDYL2 cells treated with a miR124-3p mimic or miR control (L) or in MDA-MB-231 cells co-treated with esiCDYL2 and either an anti-miR-124-3p oligonucleotide or a control anti-miR (M). Data are representative of three independent experiments.