Abstract
Background
Previous reports have demonstrated that gene expression profiles are modified in several tissue types including liver during the aging process and that such alterations might impact on the development and advancement of certain diseases. To understand the affect of aging on liver cancer, we have studied the expression pattern of cancer-related genes in aging mouse livers. Retinoid x receptor α (RXRα) is a transcription factor that is highly expressed in the liver. RXRα-regulated pathways are implicated in liver detoxification and carcinogenesis. In addition, liver cancer incidence is higher in males than in females. Thus, we have also studied the effects of hepatic RXRα and gender on regulating those cancer-related genes in aging mouse livers.
Results
Hepatic cancer-related gene expression patterns were compared between old (24-month-old) and young mature (6-month-old) wild type as well as hepatocyte RXRα-deficient C57BL/6J mice in both genders. cDNA microarray was performed and quantitative reverse transcription-polymerase chain reaction assays were employed to validate part of our results. In aged male mouse liver, genes associated with apoptosis, metastasis, cell cycle transition, and DNA repair showed changes in expression levels. In female aged mouse liver, the most notable variations were found in genes that are related to the immune system. In addition, the expression pattern is RXRα dependent.
Conclusions
Our results suggested that specific sets of genes, which are regulated by gender and RXRα, might contribute to the increase in liver cancer incidence observed with age.
Keywords: liver cancer, microarray analysis, RXRα KO mice
Introduction
Aging is defined as a genetic physiological process associated with morphological and functional changes in cellular and extra-cellular components resulting in a progressive imbalance of the control regulatory systems of the organism [1]. Cancer is the result of deregulation of gene expression and has the ability to initiate drastic change in gene expression profiles when compared to derived normal tissue. Liver cancer is one of the most prevalent cancers worldwide and its incidence rises with age. In the age group 45–49, liver cancer incidence is 10.5 and 2.1 in male and female population (100,000), respectively; while in the group of 80–84 age, the incidence rises up to 42.1 and 20.4, respectively (SEER Cancer Statistics Review, 2003, NCI). Thus, aging and being male are risk factors for liver cancer.
RXRs (Retinoid X Receptors), belonging to the nuclear receptor superfamily, play important roles in detoxification, apoptosis, differentiation, and proliferation through dimerizing with other nuclear receptors [2]. RXRα is one of the most prevalent nuclear receptors expressed in liver. Aberrant RXRα-induced pathways have been implicated as possible mechanisms for the development of hepatocellular carcinoma [3, 4]. Shortened hepatocyte lifespan and impaired capacity for liver regeneration after partial hepatectomy are detected in hepatocyte RXRα-deficient mice (RXRα KO) [5]. These findings indicate that hepatocyte RXRα also controls hepatocyte proliferation and survival.
Microarray technology has been extensively used for gene expression profile comparisons. Using this method, it has been confirmed that there are significant differences in gene expression between aged and young tissues in colon, brain, stomach, and prostate [6–8]. In these studies, the principle changes in gene expression are those related to inflammation, cellular stress, and abnormal regulation of apoptosis, cell cycle, and DNA replication. However, the mechanism of aging in each tissue is unique. It is not clear whether such differences in gene expression profiles account for aging-related diseases such as liver cancer.
In order to explore the possible molecular mechanisms that might account for increased liver cancer incidence due to aging and to understand the impact of gender and RXRα on liver cancer incidence, we performed microarray using mouse livers derived from wild type (WT) and hepatocyte RXRα-deficient mice in two age groups (6- and 24-month-old) including both genders. It is generally recognized that 6 month old mice are considered mature while 24 month old mice are aged [9, 10]. The data were analyzed based on four comparison groups: (1) WT male mice 24 vs. 6 month; (2) WT female mice 24 vs. 6 month; (3) RXRα KO male mice 24 vs. 6 month; and (4) RXRα KO female mice 24 vs. 6 month. Many genes with significant changes in mRNA levels are associated with carcinogenesis and defined as cancer-related genes by Ingenuity Pathway Analysis. We further classified these genes into the following five categories: (1) apoptosis-related genes; (2) stress-inducible related genes; (3) cell migration-related genes; (4) cell growth regulation-related genes; and (5) immune response-related genes. Our results showed that significant changes in hepatic gene expression profiles exist in the two genotypic aging mice groups, and alterations in gene expression were gender dependent. These alterations in gene expression profiles might explain the increase of liver cancer incidence in later stages of life and the interference of RXRα-mediated signaling pathways might contribute to liver carcinogenesis.
Results and Discussion
We have compared the hepatic gene expression profiles of 24-month-old mice with 6-month-old mice (WT and RXRα KO strain) in both genders. The results indicated that alterations of gene expression in aged liver are RXRα and gender dependent. For example, the top five disorders with the most significant changes in gene expression in aged livers were different in all four groups (Table 1). In aged male WT and RXRα KO mice, the number of cancer-related genes with altered mRNA levels was 202 and 123, respectively, while in aged female WT or RXRα KO mice, the number was 85 and 123, respectively (Table 2). Thus, the numbers of genes with changed expression levels due to aging were significantly less in female than in male mouse livers.
Table 1:
Top 5 categories that have the most significant difference in gene expression due to aging (24 month vs. 6 month).
| male WT | female WT | ||||
|---|---|---|---|---|---|
| disorder | Numbera | p valueb | disorder | numbera | p valueb |
| cancer | 202 | 8.41E-09 | metabolism | 28 | 1.24E-06 |
| connective tissue | 67 | 1.70E-06 | gastrointestinal | 21 | 1.99E-06 |
| development | 54 | 3.57E-06 | cancer | 85 | 2.36E-06 |
| endocrine system | 40 | 3.69E-05 | genetic | 34 | 7.50E-05 |
| metabolism | 38 | 4.61E-05 | development | 21 | 9.62E-05 |
| male KO | female KO | ||||
| disorder | numbera | p valueb | disorder | numbera | p valueb |
| cancer | 123 | 1.77E-09 | immulogical | 91 | 2.34E-22 |
| reproductive system | 57 | 2.51E-09 | connective tissue | 64 | 4.85E-20 |
| connective tissue | 36 | 2.92E-08 | inflammatory | 87 | 4.85E-20 |
| inflammatory | 55 | 2.92E-08 | skeletal and muscular | 68 | 4.85E-20 |
| skeletal and muscular | 40 | 2.92E-08 | metabolic | 49 | 4.85E-20 |
number for genes involved in disorders with at least 2 fold change in mRNA levels.
statistics significance in comparisons between 24 and 6-month-old mice.
Table 2:
Number of cancer-related genes in four aging groups.
| Male WT 24 vs. 6 month 202 genes (A+C) |
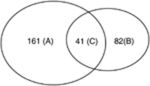
|
Male hepatocyte RXRα-deficient mice 24 vs. 6 month 123 genes (B+C) |
| Female WT 24 vs. 6 month 85 genes (A+C) |
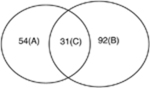
|
Female hepatocyte RXRα-deficient mice 24 vs. 6 month 123 genes (B+C) |
To validate the microarray results, we selected nine genes for quantification of mRNA levels by qRT-PCR (quantitative Reverse Transcriptase-Polymerase Chain Reaction). All qRT-PCR analyses were performed on the same samples used for the microarray experiments. Table 3 summarized the fold change in mRNA levels detected by qRT-PCR and microarray experiments. We found that both methods detected similar change trends for the up- or down-regulated genes selected for validation, although the fold changes may not be the same. The variance in fold change may reflect the difference in sensitivity of these two methods. Generally, the results from both methods are consistent.
Table 3:
Validation the change of mRNA levels in selected genes.
| male | female | ||||
|---|---|---|---|---|---|
| WT | KO | WT | KO | ||
| GADD45A | |||||
| −2.64 | −3.53 | (-) | (-) | qRT-PCR | |
| GADD45G | |||||
| −3.59 | −2.24 | (-) | (-) | qRT-PCR | |
| ATF3 | |||||
| −50.25 | (-) | (-) | (-) | qRT-PCR | |
| DNAJB1 | |||||
| −53.66 | −5.37 | (-) | (-) | qRT-PCR | |
| EGR1 | |||||
| −33.49 | (-) | (-) | (-) | qRT-PCR | |
| SERPINE1 | |||||
| −17.94 | (-) | (-) | (-) | qRT-PCR | |
| THBS1 | |||||
| −8.87 | (-) | (-) | (-) | qRT-PCR | |
| JUN | |||||
| −3.65 | (-) | (-) | (-) | qRT-PCR | |
| BTG2 | |||||
| −17.88 | (-) | (-) | (-) | qRT-PCR |
No significant change in mRNA expression level.
Cancer incidence increases during aging. In cancer cells, many encoded proteins may have abnormal function because of alterations in gene expression levels, resulting in compromised biological processes. These processes include resistance to apoptosis, unlimited replication potential, self-sufficient growth signal, insensitivity to negative regulators, sustained angiogenesis, and impaired tissue remodeling all which influence cancer cells to metastasize [11]. In addition, cell-host interactions such as immune response and cell-environment interactions such as stress response pathways have been demonstrated to play important roles in carcinogenesis [12, 13]. Thus, we classified cancer-related genes into the following five categories: apoptosis, migration, cell growth regulation, stress response and immune response, and studied their expression patterns in aging livers. Those genes whose expression levels changed more than 2-fold are summarized in each table.
Apoptosis-related genes
In aging WT male mouse liver, most pro-apoptosis genes displayed increased mRNA levels, while anti-apoptosis genes showed decreased mRNA levels in aged mouse liver. In contrast to WT male mouse liver, such patterns were not observed in the RXRα-deficient mouse livers. For example, the mRNA levels of pro-apoptosis genes in the Bcl2 family including BNIP2 (BCL2/adenovirus E1B 19KDa interacting protein 2), BIK (BCL2-interacting killer), and BID (BH3 interacting domain death agonist) were up-regulated 2.70-, 2.59-, and 2.60-fold, respectively in aged WT male mouse livers. Another pro-apoptosis gene, VDAC1 (voltage-dependent anion channel 1), also had an increase in mRNA level by 2.37-fold in aged male mouse liver. VDAC1, which is a component for the mitochondria permeability transition pore (MPT), can be modified by Bak (Bcl-2 homologous antagonist/killer) to release cytochrome c into the cytoplasm and induce apoptosis. The apoptosis effecter DFFB (DNA fragmentation factor, 40kDa), which is responsible for cutting genomic DNA into smaller fragments, was up-regulated by 2.69-fold in aged mouse liver. Conversely, the anti-apoptosis gene MCL1 (myeloid cell leukemia sequence1) showed a decrease in mRNA level by 2.59-fold in aged male mouse liver. Our data implied that the basal level of apoptosis might be higher in aged than in matured mouse livers due to changes in the expression levels of apoptosis-related genes. This finding was in agreement with other reports. Muskhelishvili et al., have demonstrated that in aged liver tissue, the rate of spontaneous apoptotic cell death was found to be elevated [14]. Other studies have shown that the expression levels of the pro-apoptotic gene gadd153/chop and DNase gamma (apoptosis-related endogenous) were elevated in aged liver [15, 16]. Recently, Kujoth et al. confirmed that the cleaved form of caspase 3 increased by 50% or higher when comparing 30- with 5- month-old liver tissue [17]. In addition to liver tissue, elevated caspase-3, −6, and −7 activity was noted in aged rat lung and spleen [18]. Our novel findings, which show a panel of genes with coordinated changes in expression patterns, provide a mechanistic explanation for the increased apoptosis observed in aging mouse liver.
Aging-associated increase of apoptosis is most likely due to a dynamic imbalance in basal levels between anti-apoptotic and pro-apoptotic genes. The Bcl2 family is one of the most important apoptosis regulators. Our data showed multiple Bcl2 family genes with altered mRNA levels and implied that the Bcl2 family might play a vital role in heightened apoptosis in aged liver. Increased apoptosis levels in aged liver might reflect a physiological response to increased DNA/protein damage due to accumulation of oxygen stress during aging, thus providing a protective mechanism to eliminate damaged cells and to maintain the whole organism in a relatively healthy condition. In aged male RXRα KO mouse livers, only a few genes (3) exhibited mRNA levels greater than 2-fold, implying that RXRα deficiency had a significant impact on the expression profile of apoptosis-related genes during aging (Table 4).
Table 4:
Fold changes of the mRNA levels of the apoptosis-related genes in aging male mouse liver.
| pro-apoptosis | fold (24 vs. 6 month) | ||
|---|---|---|---|
| Name | Description | WT | RXRα deficiency |
| SNCA | synuclein, alpha | 5.37 | (-) |
| ACVR2B | activin A receptor, type IIB | 3.91 | (-) |
| BNIP2 | BCL2/adenovirus E1B 19kDa interacting protein 2 | 2.72 | (-) |
| DFFB | DNA fragmentation factor, 40kDa, | 2.69 | (-) |
| BID | BH3 interacting domain death agonist | 2.61 | (-) |
| BIK | BCL2-interacting killer (apoptosis-inducing) | 2.59 | (-) |
| VDAC1 | voltage-dependent anion channel 1 | 2.37 | (-) |
| DAPK1 | death-associated protein kinase 1 | 2.27 | (-) |
| TFAP4 | transcription factor AP-4 | 2.25 | (-) |
| PDCD6IP | programmed cell death 6 interacting protein | −2.38 | (-) |
| BCL2L11 | BCL2-like 11 (bim) | −4.46 | (-) |
| SOX4 | SRY-box containing gene 4 | (-) | 2.31 |
| anti-apoptosis | fold (24 vs. 6 month) | ||
| Name | Description | WT | RXRα deficiency |
| PHLDA1 | pleckstrin homology-like domain, family A, | −2.33 | (-) |
| MCL1 | myeloid cell leukemia sequence 1 (BCL2-related) | −2.59 | (-) |
| IER3 | immediate early response 3 | −2.62 | (-) |
| BCL2L1 | Bcl2-like 1 (BCL-XL) | (-) | −2.35 |
| USP2 | ubiquitin specific protease 2 | (-) | 3.07 |
Stress response-related genes
Many HSP (Heat Shock Protein) genes had decreased mRNA levels in aged male WT and RXRα KO livers (Table 5). The HSP family is highly conserved structurally from C. elegans to human. HSP genes constitute the cellular protection mechanism and can be induced by various physical, chemical, and biological stimuli. Our data showed that HSP1A (Heat Shock 70 KDa Protein 1A), HSP1B (Heat Shock 70 KDa Protein 1B), and DNAJB1 (Dnaj homologue, subfamily B, member 1) mRNA levels decreased by 44.84-, 28.01-, and 27.10-fold, in WT male mice, respectively. In RXRα KO mouse livers, the fold reductions were not so high; the reduction was below threshold for HSP1A, 3.94 for HSP1B, and 2.13 for DNAJB1. Conflicting findings have been cited with regard to the HSP family basal expression levels in various tissues with age. In aged peripheral blood mononuclear cell and rat kidney, the mRNA level of HSP70 can be found decreased or increased, respectively [19, 20]. In senescent fibroblast cells, the HSP47 level was found to be decreased [21] while in brain tissue, the level of HSP72 and HSP70 mRNA was found to increase with age [22]. In liver, the basal expression level of HSP25 was reported to increase in aging C3B10RF1 mice [10]. Thus, it seems that the alteration trends in basal expression levels for HSP family genes might be tissue or species specific.
Table 5:
Fold changes of the mRNA levels of the stress inducible-genes in aging male mouse liver.
| non-genotoxic stress-inducible genes | Fold change (24 vs. 6 month) | ||
|---|---|---|---|
| Name | Description | WT | RXRα deficiency |
| HSPA8 | heat shock 70kDa protein 8 | −2.04 | (-) |
| HSP90B1 | heat shock protein 90kDa beta, member 1 | −2.39 | −2.19 |
| DNAJB4 | DnaJ (Hsp40) subfamily B, member 4 | −2.40 | (-) |
| HYOU1 | hypoxia up-regulated 1 | −2.56 | −3.65 |
| HSPB8 | heat shock 22kDa protein 8 | −3.22 | −2.11 |
| HSP90AA1 | heat shock protein 90kDa alpha | −4.79 | −3.18 |
| HSPB1 | heat shock 27kDa protein 1 | −6.41 | −4.91 |
| DNAJB1 | DnaJ (Hsp40) subfamily B, member 1 | −27.10 | −2.13 |
| HSPA1B | heat shock 70kDa protein 1B | −28.01 | −3.94 |
| HSPA1A | heat shock 70kDa protein 1A | −44.84 | (-) |
| genotoxic stress-inducible genes | Fold change (24 vs. 6 month) | ||
| Name | Description | WT | RXRα deficiency |
| OSGIN1 | oxidative stress induced growth inhibitor 1 | 3.24 | 2.10 |
| DDIT3 | DNA-damage-inducible transcript 3 | −2.24 | (-) |
| GADD45G | growth arrest and DNA-damage, gamma | −3.61 | −2.91 |
| GADD45A | growth arrest and DNA-damage alpha | −4.43 | −4.91 |
| ATF3 | activating transcription factor 3 | −33.01 | (-) |
There is a general tendency of delayed or decreased induction expression level for HSP family genes in aging tissues [23, 24]. Our data showed that multiple HSP genes had decreased mRNA levels in both aging WT and RXRα KO male mice livers although fold changes in mRNA levels were very different. This coordinated down-regulation of basal levels in HSP family genes might contribute to a delayed response or reduced induction level to exogenous stimuli in aging livers. HSP family genes are chaperons that ensure proper folding and normal function of proteins and thus provide a protective effect against non-genotoxic stimulation such as increased environmental temperature. Aged cells have higher levels of damaged proteins or lipids because of reactive oxygen species accumulation. The down-regulation of the HSP family gene expression levels implies that aged cells might not have sufficient protection and thus might undergo apoptosis, providing another possible explanation for the observed increased basal apoptosis in aged liver.
Other types of stress-induced (DNA damaged induced) genes which were also down-regulated include ATF3 (Activating Transcription Factor 3, 33.00-fold down), GADD45 family (Growth Arrested and DNA Damage induced alpha and gamma, 4.42- and 3.61-fold down, respectively), and DDIT3 (DNA Damage Inducible Transcription 1, 2.24-fold down). In RXRα KO mice, ATF3 and DDIT3 showed no change in mRNA levels, while GADD45G and GADD45A mRNA levels were reduced by 2.91- and 4.91-fold, respectively. In response to genotoxic stimuli, the mRNA levels of these genes are highly induced and consequently lead to apoptosis. ATF3 has been demonstrated to increase the transcription of p53 in response to stimuli, and the GADD family is the downstream effector of the p53 pathway. Thus, our data suggested that the p53 signaling pathway is inhibited in aged male mouse liver although p53 mRNA level was not changed. This finding is consistent with a previous report which showed the impairment of the p53-GADD45 signaling pathway in aged heart [24].
In aged rat liver tissue, the number of genotoxic compound-induced apoptotic cells is significantly lower than that found in young rat liver [25]. It was believed that in aged tissues, the signaling systems known to be involved in genotoxic stress-induced apoptosis become deregulated with age [25]. Because down-regulation of p53 signaling pathway leads to a reduction of apoptosis of DNA damaged cells, we conclude it is very likely that reduced genotoxic compound-induced apoptosis in aging WT liver results from a compromised p53 signaling pathway. In male RXRα KO mice, the fold changes in expression levels for stress-induced genes were generally less in comparison with WT mice. For example, HSPA1A and ATF3 mRNA levels, which decreased drastically in WT, were not changed in KO aging liver. Our data clearly indicated that RXRα deficiency reduced the changes in stress-induced gene expression levels due to aging.
Cell migration-related genes
Genes in this group play important roles in cell migration and angiogenesis and are associated with metastasis, one of the most important phenotypes of malignant cancer cells. As shown in Table 6, many pro-metastasis genes depicted increased levels of mRNA, however the mRNA levels of anti-metastasis genes were reduced in aging WT male mouse liver. Generally, in RXRα-deficient liver, fewer genes showed changes in mRNA level. Only four (BGN, CD36, DHCR24, and CXCL10) pro-metastasis genes had elevated mRNA levels and one (NDG1) anti-metastasis gene showed decreased mRNA levels. The MMP (Matrix Metallopeptidase) family, which degrades the extra-cellular matrix and promotes cancer cell migration, plays critical roles in metastasis [26]. The mRNA level of MMP14 increased 2.10-fold, while the level of TIMP2 and TIMP3 (Tissue Metallopeptidase Inhibitor 2, 3) decreased 2.34- and 2.12-fold, respectively. This finding implies that in aged mouse WT liver, cell-matrix interaction is in a relatively loose state, favoring cell migration.
Table 6:
Fold changes of the mRNA levels of cell migration-related genes in aging male mouse liver.
| anti-metastasis-related genes | Fold change (24 vs. 6 month) | ||
|---|---|---|---|
| Name | Description | WT | RXRα deficiency |
| TIMP3 | TIMP metallopeptidase inhibitor 3 | −2.12 | (-) |
| TIMP2 | TIMP metallopeptidase inhibitor 2 | −2.35 | (-) |
| VCL | vinculin | −2.68 | (-) |
| MSN | moesin | −3.18 | (-) |
| ITGAM | integrin, alpha M | −3.51 | (-) |
| CTGF | connective tissue growth factor | −4.53 | (-) |
| THBS1 | thrombospondin 1 | −19.53 | (-) |
| SERPINE1 | serpin peptidase inhibitor, clade E | −20.49 | (-) |
| NDG1 | N-myc downstream regulated gene 1 | (-) | −2.66 |
| pro-metastasis-related genes | Fold change (24 vs. 6 month) | ||
| Name | Description | WT | RXRα deficiency |
| JUB | jub, ajuba homolog (Xenopus laevis) | 4.58 | (-) |
| CTTN | cortactin | 3.64 | (-) |
| FN1 | fibronectin 1 | 2.96 | (-) |
| TPM3 | tropomyosin 3 | 2.82 | (-) |
| LPP | LIM domain | 2.6 | (-) |
| DAG1 | dystroglycan 1 | 2.53 | (-) |
| DDR1 | discoidin domain receptor family,1 | 2.37 | (-) |
| DSG2 | desmoglein 2 | 2.27 | (-) |
| PTP4A3 | PTP type IV member 3 | 2.11 | (-) |
| MMP14 | matrix metallopeptidase 14 | 2.11 | (-) |
| VIM | vimentin | −2.21 | (-) |
| PLAUR | plasminogen activator | −3.26 | (-) |
| NEDD9 | neural precursor cell expressed, | −4.02 | (-) |
| BGN | biglycan | (-) | 2.06 |
| CD36 | CD36 | (-) | 2.32 |
| DHCR24 | 24-dehydrocholesterol reductase | (-) | 2.63 |
| CXCL10 | CXCL10 | (-) | 2.72 |
The mRNA levels of THBS1 (Thrombospondin 1, 19.53-fold) and SERPINE (Serpin Peptidase Inhibitor, 20.49-fold) also demonstrated a striking down-regulation in aged WT male mice livers. THBS1 selectively inhibits early-stage carcinogenesis and angiogenesis in skin cancer [27]. SERPINE1, also named PAI-1 (plasminogen activation inhibitor 1), has been used in gene therapy for inhibition of melanoma metastasis [28]. The expression levels of pro-metastasis genes including JUB (Jub, 4.59-fold) and CTTN (Cortactin, 3.64-fold) were up-regulated. JUB and CTTN modulate cytoskeleton structure and increase cell movement [29, 30]. Taken together, such alterations in cell migration-related gene expression profiles occurring in aged male WT mouse liver would impair cell-cell and cell-matrix interactions and lead to the alteration of micro-environments promoting cell migration. As for RXRα KO mice, no appreciable changes in migration-related gene expression levels have been identified. Our results implied that RXRα deficiency could also reduce the variation in expression of migration-related genes due to aging.
Cell growth regulation-related genes
In comparison with the other categories, the cell growth regulation-related gene category has the highest number of genes that demonstrate changed mRNA levels in aged WT male mouse liver (Table 7). Both the positive and negative cell growth regulators had altered mRNA levels. For example, the mRNA levels of proto-oncogenes Jun, Fos, and Myc were decreased by 6.53-, 7.14-, and 17.07-fold, respectively. It has been well established that the proliferative potential of aged livers declines and that the down-regulation of Myc is the key event responsible for this decrease [31]. Down-regulation of Fos, an AP-1 transcription factor, was found in aged rat liver [32]. Inhibition of EGF (Epidermal Growth Factor) signaling pathway was also noted in aged rat liver [33]. Thus, our findings are consistent with those data.
Table 7:
Fold changes of the mRNA levels of cell growth regulation-related genes in aging male mouse liver.
| cell growth-inhibition related genes | Fold change (24 vs. 6 month) | ||
|---|---|---|---|
| Name | Description | WT | RXRα deficiency |
| SMARCB1 | SWI/SNF related,member 1 | 2.62 | (-) |
| BACH2 | basic leucine zipper transcription actor2 | 2.38 | (-) |
| DLEU2 | deleted in lymphocytic leukemia, 2 | 2.28 | (-) |
| RNF6 | ring finger protein (C3H2C3 type) 6 | 2.27 | (-) |
| RASSF5 | Ras association domain family 5 | 2.27 | (-) |
| ING2 | inhibitor of growth family, member 2 | 2.22 | (-) |
| LATS2 | LATS, large tumor suppressor, | 2.02 | (-) |
| SOD1 | superoxide dismutase 1, soluble | 2.01 | (-) |
| CEACAM1 | Carcinoembryonic cell adhesion molecule | −2.05 | (-) |
| EDNRB | endothelin receptor type B | −2.07 | (-) |
| SEL1L | sel-1 suppressor of lin-12-like | −2.08 | (-) |
| LASP1 | LIM and SH3 protein 1 | −2.08 | (-) |
| JDP2 | jun dimerization protein 2 | −2.16 | (-) |
| ID4 | inhibitor of DNA binding 4, | −2.34 | (-) |
| RHOB | ras homolog gene family, member B | −2.72 | (-) |
| FLNA | filamin A, alpha | −2.93 | (-) |
| TAGLN | transgelin | −3.02 | (-) |
| CDKN1A | p21, Cip1 | −4.65 | (-) |
| DUSP6 | dual specificity phosphatase 6 | −4.65 | (-) |
| PLK3 | polo-like kinase 3 (Drosophila) | −5.26 | (-) |
| KLF6 | Kruppel-like factor 6 | −6.9 | (-) |
| DUSP1 | dual specificity phosphatase 1 | −7.41 | (-) |
| EGR2 | early growth response 2 | −7.46 | (-) |
| BTG2 | BTG family, member 2 | −9.35 | (-) |
| EGR1 | early growth response 1 | −13.48 | (-) |
| WEE1 | Wee1 | (-) | 5.23 |
| GPX3 | glutathione peroxidase 3 | (-) | 3.78 |
| SOCS2 | suppressor of cytokine signaling 2 | (-) | 3.02 |
| FHIT | fragile histidine triad gene pleckstrin homology domain interacting | (-) | 2.67 |
| PHIP | protein | (-) | 2.17 |
| DEFB1 | defensin, beta 1 | (-) | 2.02 |
| NFIL3 | nuclear factor, interleukin 3, regulated | (-) | −2.47 |
| SOCS3 | suppressor of cytokine signaling 3 | (-) | −2.91 |
| cell growth-stimulation related genes | Fold change (24 vs. 6 month) | ||
| Name | Description | WT | RXRα deficiency |
| TFAP2A | transcription factor AP-2 alpha | 2.16 | (-) |
| PIM1 | pim-1 oncogene | 2.06 | (-) |
| MDM2 | Mdm2, p53 binding protein (mouse) | −2.05 | (-) |
| TFF3 | trefoil factor 3 (intestinal) | −2.05 | (-) |
| MAP3K14 | mitogen-activated protein 3 kinase 14 | −2.21 | (-) |
| FYN | FYN oncogene | −2.36 | (-) |
| CRKL | v-crk sarcoma virus CT10 oncogene | −2.53 | (-) |
| AXL | AXL receptor tyrosine kinase | −2.65 | (-) |
| CXCL2 | chemokine (C-X-C motif) ligand 2 | −2.67 | (-) |
| SERTAD1 | SERTA domain containing 1 | −2.71 | (-) |
| SPHK1 | sphingosine kinase 1 | −2.79 | (-) |
| PTTG1 | pituitary tumor-transforming 1 | −2.95 | (-) |
| ETS2 | v-ets oncogene | −3.45 | (-) |
| JUNB | jun B proto-oncogene | −3.77 | (-) |
| JUN | jun oncogene | −6.54 | (-) |
| FOS | v-fos oncogene | −7.14 | (-) |
| MYC | v-myc oncogene | −17.07 | (-) |
| CD74 | CD74 | (-) | 2.16 |
| CRIP1 | cysteine-rich protein 1 | (-) | 2.31 |
| PML | promyelocytic leukemia | (-) | 5.38 |
| PROM1 | prominin 1 | (-) | 2.63 |
| RBM3 | RNA binding motif protein 3 | (-) | 3.35 |
| S100A6 | S100 calcium binding protein A6 | (-) | 5.81 |
| SLPI | leukocyte peptidase inhibitor | (-) | 3.39 |
Our data showed that down-regulation of cell growth negative regulators occurred in aged male WT mouse livers. Tumor suppressor genes including p21 (4.65-fold), KLF6 (Kruppel-like factor 6, 6.90-fold), BTG2 (B-cell Translocation Gene-2, 9.35-fold), and EGR1 and 2 (Early Growth Response 1 and 2, 13.48- and 7.46-fold, respectively) were all down-regulated in aged male mouse liver. The mRNA levels of p21 are lowered in aged rat hepatocytes [34], which is consistent with our result (4.65-fold down). One of the p21 target genes, KLF6, also showed a decrease in mRNA level. Another tumor suppressor gene, BTG2, is closely associated with aging and cancer [35] and is a major downstream target of the p53 pathway [36]. BTG2 functions as a cell cycle inhibitor and its mRNA level decreased by 9.35 fold. Many negative regulators of the cell cycle exhibited changes in mRNA levels implying cell cycle checkpoint machinery might be deregulated in aged male mouse liver. The impairment of negative controls for cell cycling would increase the possibility of indefinite cell growth and elevate the risk for cancer development.
Among the five categories of cancer related genes, the cell growth regulation category has the highest number of genes with altered mRNA levels in KO aging mouse livers. However, contrary to WT mouse liver, most of the cell-growth inhibition genes and all the cell-growth stimulation genes showed elevated mRNA levels in RXRα KO aging liver. Remarkably, there was no gene which depicted changed mRNA levels both in WT and RXRα KO aging livers. For example, JUN, FOS, and MYC all exhibited decreased expression levels in WT aging mice, but such changes were not found in RXRα KO aging mouse liver at all. Our results indicated RXRα deficiency had a significant impact on aging-associated liver cell growth and proliferation.
Immune response-related genes
In contrast to the changed genetic profiles in aging male mouse livers, the above-described trends were not observed in female mouse liver. The most striking changes that occurred in aged female mouse liver were found in immune response-related genes (Table 8). For example, C1QA (complement component 1), IGK (immunoglobulin kappa chain), and IGHM (immunoglobulin heavy constant mu) mRNA levels were increased by 3.94-, 3.72-, and 4.03-fold, respectively. The induction of these genes suggests the presence of chronic inflammation in aged female mouse livers. This finding was consistent with previous reports, which showed that aging is often associated with de-regulation of the immune response even among healthy elderly women [37]. Chronic inflammation is closely linked with tumorigenesis [38]. Our results suggest that inflammation might be a key component in the increased liver cancer incidence observed in aging female mice. Thus, the mechanisms that increase the risk for liver cancer due to aging might be different between male and female mice.
Table 8:
Fold changes for the mRNA level of immune response-related genes in aging female liver.
| immune-response related genes | Fold change (24 vs. 6 month) | ||
|---|---|---|---|
| Name | Description | WT | RXRα deficiency |
| PTPRC | PTP, receptor type, C | 2.31 | 4.08 |
| CD52 | CD52 molecule | 2.38 | 3.27 |
| CTSS | cathepsin S | 2.48 | 3.03 |
| TYROBP | TYRO protein binding protein | 2.53 | (-) |
| TGTP | T-cell specific GTPase | 2.85 | (-) |
| CD74 | CD74, major histocompatibility complex | 2.93 | 3.32 |
| LILRB4 | leukocyte immunoglobulin-like receptor, | 3.31 | (-) |
| IGK | immunoglobulin kappa chain complex | 3.72 | (-) |
| C1QA | complement component 1 | 3.94 | (-) |
| IGHM | immunoglobulin heavy constant mu | 2.91 | 5.25 |
| IGHG1 | immunoglobulin heavy constant gamma 1 | 4.29 | 7.83 |
| HLA-DMA | major histocompatibility complex, class II, | (-) | 2.24 |
| CXCL9 | chemokine (C-X-C motif) ligand 9 | (-) | 2.41 |
| IL1B | interleukin 1, beta | (-) | 2.60 |
| CXCL14 | chemokine (C-X-C motif) ligand 14 | (-) | 3.43 |
| IL2RG | interleukin 2 receptor, gamma | (-) | 5.00 |
| IGH-1A | immunoglobulin heavy chain 1a | (-) | 48.56 |
Immune response-related genes had the same change trend in RXRα KO aging female mice. Six genes showed increased mRNA levels in both WT and RXRα KO strains and the folds were all higher in RXRα KO than in WT aging liver. Furthermore, IGH-1A (immunoglobulin heavy chain 1a) had increased expression level by 48.56-fold while no change occurred in WT aging liver. Our data indicated that RXRα deficiency promotes the change trend of immune-response related genes due to aging in female mice. Our findings are consistent with previous reports that RXRα deficiency increases inflammation response to stimulus, which might be due to up regulation of a panel of immune-related genes [39].
Summary:
By comparing the gene expression patterns between aged and young liver tissues in both WT and RXRα KO strains, our data indicated that genes, which regulate cell apoptosis, damage response, migration, and growth, are de-regulated in aging male WT mouse livers. There is a strong positive association between aging and cancer incidence, but the molecular mechanism is unclear. Cumulative cellular damage contributes to both aging and cancer. However, telomerase shortening and cell cycle checkpoint machinery such as the p53 pathway can prevent cancer predisposition but promote aging processes [40]. Therefore, the mechanisms involved in aging and cancer are intertwined. The alteration in gene expression in aging liver might explain the relationship between aging and increased incidence of liver cancer.
The change in the expression level of cancer-related genes in aging mouse liver is gender specific. In aging female WT mice, in contrast to aging male liver, the major pathway altered was the immune-related pathway. It is well known that female hormones play a critical role in maintaining or enhancing immune responses [41]. Hormonal changes that lead to altered immune response might play a very important role in age-associated increase of liver cancer observed in females. There still may be other genes in different pathways that are also important in age-associated increase or gender-associated difference of liver cancer incidence.
The role of hepatocyte RXRα has been well characterized [42–46]. Hepatocyte RXRα not only regulates endobiotic and xenobiotic metabolism, it also dimerizes with other nuclear receptors and plays an important role in inflammation process and liver regeneration [39, 47]. Hepatocyte RXRα deficiency has various impacts on different categories of cancer-related genes in aging male and female liver. RXRα deficiency reduced the number of apoptosis-related and migration-related genes with altered mRNA levels due to aging. Most stress-inducible genes showed the same down-regulation trend in expression levels in both WT and RXRα KO strains but fewer fold changes were observed in RXRα KO mice. RXRα deficiency gave rise to increased expression levels of cell growth-related genes in KO mouse livers. In female mice, RXRα deficiency aggravated the up-regulation of immune-response related genes, which implies increased immune response sensitivity existed in aging female mouse liver. Overall, RXRα deficiency affects the expression profile of cancer-related genes due to aging in male and female mice. It is very likely that RXRα deficiency has some effects on increased incidence of liver cancer due to aging.
Conclusions
Significant changes in hepatic gene expression profiles occurred during aging in both genders. These changes might increase the susceptibility of aged mice to liver cancer. RXRα deficiency affects the age-associated changes of hepatic expression profiles, which may affect the increased liver cancer incidence observed with age.
Materials and methods
Animals
Animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Kansas Medical Center, Kansas City. Male and Female C57BL/6J mice were weaned at 28 days, housed individually, given free access to water, and randomly assigned to study groups. Four groups of mice were used to determine the effects of aging in both male and female mice. Each group had 3 mice. At particular time points after birth, 6 months (mature) and 24 months (aged) mice were sacrificed by cervical dislocation, and the livers were rapidly excised and flash frozen in liquid nitrogen. No abnormal pathology was detected in any of the animals used.
RNA Isolation and Preparation for Microarray Hybridization
Total RNA was isolated from frozen mouse liver using Trizol Reagent (Invitrogen Corporation, Carlsbad, CA) and further purified using the RNeasy Mini Kit (Qiagen Inc., Valencia, CA). Total RNA was quantified by UV spectrophotometry and its integrity and quality were assessed on RNA 6000 Nano LabChips with the Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA). Synthesis and purification of double-stranded cDNA were conducted as described in the Expression Analysis Technical Manual (Affymetrix, Santa Clara, CA). Biotin-labeled cRNA was synthesized from the purified cDNA using the BioArray High Yield Transcript Labeling Kit according to the manufacturer’s specifications (ENZO Life Sciences, Farmingdale, NY). Labeled cRNA was purified using the GeneChip Sample Cleanup Module, quantified by UV spectrophotometry and assessed for quality with the Bioanalyzer 2100. Twenty μg purified cRNA was fragmented and 15 μg fragmented cRNA was hybridized to Affymetrix Mouse Genome 430A 2.0 Array GeneChips (Affymetrix, Santa Clara, CA) according to the Expression Analysis Technical Manual. Washing and staining of the hybridized arrays were carried out using the Fluidics Station 400 and arrays were subsequently scanned with the Hewlett Packard GeneArray Scanner.
Microarray Data Analysis
Affymetrix scan data (.cel files) were imported into Rosetta Resolver for analysis (Rosetta Biosoftware, Seattle, WA). Following intrachip normalization and background correction, individual replicates were combined into single “ratio experiments” by an error-weighted method using the control group as a baseline. An agglomerative hierarchical clustering algorithm utilizing an error-weighted Euclidian distance measure was performed on the ratio experiments to identify active genes. Transcripts were defined as active (with significant change) if they increased or decreased their levels by greater than 2-fold.
Confirmation of mRNA level by quantitative real-time PCR
The synthesized cDNA was diluted 20 fold in water. All primers and probes were designed based on nucleotide sequences in Genbank using the Primer Express software (PE Applied Biosystems). PCR reaction efficiency was calculated for each primer pair using with five dilution points of the calibrator sample to validate primers and probes. The PCR product covered at least two exons according to introns-exons organization of selected genes. Each real-time PCR consisted of 1x PCR Master Mix (PE Applied Biosystems), 0.5 μM forward and reverse primers and 1uM corresponding probe. Final reaction volume was 20 μl. Reactions were carried out on ABI PRISM 7900 real time PCR system (PE Applied Biosystems) for 40 cycles (95°C for 15 s, 60°C for 1 min). The expression fold change for each gene was calculated using the Ct method and beta-actin was used as an internal control.
Acknowledgement:
This work is supported by NIH grants CA53596, AA12081, AA14147, and COBRE grant P20 RR021940. We thank Ms. Barbara Brede for proofreading of this manuscript.
Footnotes
Competition interests:
The authors declare that they have no conflict of interests.
References
- [1].Finkel T, Serrano M, and Blasco MA, The common biology of cancer and ageing, Nature, 448 (2007), 767–774. [DOI] [PubMed] [Google Scholar]
- [2].Altucci L, et al. , RAR and RXR modulation in cancer and metabolic disease, Nat Rev Drug Discov, 6 (2007), 793–810. [DOI] [PubMed] [Google Scholar]
- [3].Kanamori T, et al. , Synergistic growth inhibition by acyclic retinoid and vitamin K2 in human hepatocellular carcinoma cells, Cancer Sci, 98 (2007), 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Moriwaki H, et al. , Chemoprevention of liver carcinogenesis with retinoids: Basic and clinical aspects, Hepatol Res, 37 Suppl 2 (2007), S299–302. [DOI] [PubMed] [Google Scholar]
- [5].Imai T, et al. , Selective ablation of retinoid X receptor alpha in hepatocytes impairs their lifespan and regenerative capacity, Proc Natl Acad Sci U S A, 98 (2001), 4581–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hallberg LM, et al. , Effects of aging and caloric restriction on IGF-I, IGF-I receptor, IGFBP-3 and IGFBP-4 gene expression in the rat stomach and colon, Regul Pept, 89 (2000), 37–44. [DOI] [PubMed] [Google Scholar]
- [7].Weindruch R, and Prolla TA, Gene expression profile of the aging brain, Arch Neurol, 59 (2002), 1712–1714. [DOI] [PubMed] [Google Scholar]
- [8].Reyes I, et al. , Aging-associated changes in gene expression in the ACI rat prostate: Implications for carcinogenesis, Prostate, 63 (2005), 169–86. [DOI] [PubMed] [Google Scholar]
- [9].Frick KM, et al. , Reference memory, anxiety and estrous cyclicity in C57BL/6NIA mice are affected by age and sex, Neuroscience, 95 (2000), 293–307. [DOI] [PubMed] [Google Scholar]
- [10].Cao SX, et al. , Genomic profiling of short- and long-term caloric restriction effects in the liver of aging mice, Proc Natl Acad Sci U S A, 98 (2001), 10630–10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hanahan D, and Weinberg RA, The hallmarks of cancer, Cell, 100 (2000), 57–70. [DOI] [PubMed] [Google Scholar]
- [12].Karin M, Lawrence T, and Nizet V, Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer, Cell, 124 (2006), 823–835. [DOI] [PubMed] [Google Scholar]
- [13].Harkin DP, et al. , Induction of GADD45 and JNK/SAPK-dependent apoptosis following inducible expression of BRCA1, Cell, 97 (1999), 575–586. [DOI] [PubMed] [Google Scholar]
- [14].Muskhelishvili L, et al. , Age-related changes in the intrinsic rate of apoptosis in livers of diet-restricted and ad libitum-fed B6C3F1 mice, Am J Pathol, 147 (1995), 20–24. [PMC free article] [PubMed] [Google Scholar]
- [15].Ikeyama S, et al. , Expression of the pro-apoptotic gene gadd153/chop is elevated in liver with aging and sensitizes cells to oxidant injury, J Biol Chem, 278 (2003), 16726–16731. [DOI] [PubMed] [Google Scholar]
- [16].Tanaka K, et al. , Aging increases DNase gamma, an apoptosis-related endonuclease, in rat liver nuclei: effect of dietary restriction, Exp Gerontol, 39 (2004),195–202. [DOI] [PubMed] [Google Scholar]
- [17].Kujoth GC, et al. , Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging, Science, 309 (2005), 481–484. [DOI] [PubMed] [Google Scholar]
- [18].Zhang Y, Chong E, and Herman B, Age-associated increases in the activity of multiple caspases in Fisher 344 rat organs, Exp Gerontol, 37 (2002), 777–789. [DOI] [PubMed] [Google Scholar]
- [19].Deguchi Y, Negoro S, and Kishimoto S, Age-related changes of heat shock protein gene transcription in human peripheral blood mononuclear cells, Biochem Biophys Res Commun, 157 (1988), 580–584. [DOI] [PubMed] [Google Scholar]
- [20].Maiello M, et al. , Basal synthesis of heat shock protein 70 increases with age in rat kidneys, Gerontology, 44 (1998), 15–20. [DOI] [PubMed] [Google Scholar]
- [21].Nizard C, et al. , Heat shock protein 47 expression in aged normal human fibroblasts: modulation by Salix alba extract, Ann N Y Acad Sci, 1019 (2004), 223–227. [DOI] [PubMed] [Google Scholar]
- [22].Calabrese V, et al. , Increased expression of heat shock proteins in rat brain during aging: relationship with mitochondrial function and glutathione redox state, Mech Ageing Dev, 125 (2004), 325–35. [DOI] [PubMed] [Google Scholar]
- [23].Rao DV, Watson K, and Jones GL, Age-related attenuation in the expression of the major heat shock proteins in human peripheral lymphocytes, Mech Ageing Dev, 107 (1999), 105–18. [DOI] [PubMed] [Google Scholar]
- [24].Edwards MG, et al. , Impairment of the transcriptional responses to oxidative stress in the heart of aged C57BL/6 mice, Ann N Y Acad Sci, 1019 (2004), 85–95. [DOI] [PubMed] [Google Scholar]
- [25].Suh Y, et al. , Aging alters the apoptotic response to genotoxic stress, Nat Med, 8 (2002), 3–4. [DOI] [PubMed] [Google Scholar]
- [26].Arai I, et al. , Overexpression of MT3-MMP in hepatocellular carcinoma correlates with capsular invasion, Hepatogastroenterology, 54 (2007), 167–171. [PubMed] [Google Scholar]
- [27].Hawighorst T, et al. , Thrombospondin-1 selectively inhibits early-stage carcinogenesis and angiogenesis but not tumor lymphangiogenesis and lymphatic metastasis in transgenic mice, Oncogene, 21 (2002), 7945–7956. [DOI] [PubMed] [Google Scholar]
- [28].Ma D, et al. , Inhibition of metastasis of intraocular melanomas by adenovirus-mediated gene transfer of plasminogen activator inhibitor type 1 (PAI-1) in an athymic mouse model, Blood, 90 (1997), 2738–2746. [PubMed] [Google Scholar]
- [29].Pratt SJ, et al. , The LIM protein Ajuba influences p130Cas localization and Rac1 activity during cell migration, J Cell Biol, 168 (2005), 813–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Luo ML, and Wang MR, CTTN (EMS1): an oncogene contributing to the metastasis of esophageal squamous cell carcinoma, Cell Res, 17 (2007), 298–300. [DOI] [PubMed] [Google Scholar]
- [31].Iakova P, Awad SS, and Timchenko NA, Aging reduces proliferative capacities of liver by switching pathways of C/EBPalpha growth arrest, Cell, 113 (2003), 495–506. [DOI] [PubMed] [Google Scholar]
- [32].Goyns MH, et al. , Differential display analysis of gene expression indicates that age-related changes are restricted to a small cohort of genes, Mech Ageing Dev, 101 (1998), 73–90. [DOI] [PubMed] [Google Scholar]
- [33].Ikeyama S, et al. , Effects of aging and calorie restriction of Fischer 344 rats on hepatocellular response to proliferative signals, Exp Gerontol, 38 (2003), 431–439. [DOI] [PubMed] [Google Scholar]
- [34].Kitano S, et al. , Effect of aging on regulation of sdi-1 in rat hepatocytes, Biochem Biophys Res Commun, 225 (1996), 122–127. [DOI] [PubMed] [Google Scholar]
- [35].Lim IK, TIS21 (/BTG2/PC3) as a link between ageing and cancer: cell cycle regulator and endogenous cell death molecule, J Cancer Res Clin Oncol, 132 (2006), 417–426. [DOI] [PubMed] [Google Scholar]
- [36].Boiko AD, et al. , A systematic search for downstream mediators of tumor suppressor function of p53 reveals a major role of BTG2 in suppression of Ras-induced transformation, Genes Dev, 20 (2006), 236–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ahluwalia N, Aging, nutrition and immune function, J Nutr Health Aging, 8 (2004), 2–6. [PubMed] [Google Scholar]
- [38].Balkwill F, TNF-alpha in promotion and progression of cancer, Cancer Metastasis Rev, 25 (2006), 409–416. [DOI] [PubMed] [Google Scholar]
- [39].Gyamfi MA, et al. , Hepatocyte retinoid X receptor alpha-dependent regulation of lipid homeostasis and inflammatory cytokine expression contributes to alcohol-induced liver injury, J Pharmacol Exp Ther, 324 (2008), 443–453. [DOI] [PubMed] [Google Scholar]
- [40].Serrano M, and Blasco MA, Cancer and ageing: convergent and divergent mechanisms, Nat Rev Mol Cell Biol, 2007. 8(2007), 715–22. [DOI] [PubMed] [Google Scholar]
- [41].Cutolo M, et al. , Estrogens and autoimmune diseases, Ann N Y Acad Sci, 1089 (2006), 538–47. [DOI] [PubMed] [Google Scholar]
- [42].Wan YJ, et al. , Peroxisome proliferator-activated receptor alpha-mediated pathways are altered in hepatocyte-specific retinoid X receptor alpha-deficient mice, J Biol Chem, 275 (2000), 28285–28290. [DOI] [PubMed] [Google Scholar]
- [43].Wan YJ, et al. , Hepatocyte-specific mutation establishes retinoid X receptor alpha as a heterodimeric integrator of multiple physiological processes in the liver, Mol Cell Biol, 20 (2000), 4436–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wan YJ, et al. , Hepatocyte retinoid X receptor-alpha-deficient mice have reduced food intake, increased body weight, and improved glucose tolerance, Endocrinology, 144 (2003), 605–611. [DOI] [PubMed] [Google Scholar]
- [45].Dai G, and Wan YJ, Animal models of xenobiotic receptors, Curr Drug Metab, 6 (2005), 341–55. [DOI] [PubMed] [Google Scholar]
- [46].Gyamfi MA, and Wan YJ, The effect of ethanol, ethanol metabolizing enzyme inhibitors, and Vitamin E on regulating glutathione, glutathione S-transferase, and S-adenosylmethionine in mouse primary hepatocyte, Hepatol Res, 35 (2006), 53–61. [DOI] [PubMed] [Google Scholar]
- [47].Dai G, et al. , Pregnane X receptor is essential for normal progression of liver regeneration, Hepatology, 47 (2008), 1277–87. [DOI] [PubMed] [Google Scholar]


