Abstract
With the current trajectory of the 2019-nCoV outbreak unknown, public health and medicinal measures will both be needed to contain spreading of the virus and to optimize patient outcomes. While little is known about the virus, an examination of the genome sequence shows strong homology with its more well-studied cousin, SARS-CoV. The spike protein used for host cell infection shows key nonsynonymous mutations which may hamper efficacy of previously developed therapeutics but remains a viable target for the development of biologics and macrocyclic peptides. Other key drug targets, including RdRp and 3CLpro, share a strikingly high (>95%) homology to SARS-CoV. Herein, we suggest 4 potential drug candidates (an ACE2-based peptide, remdesivir, 3CLpro-1 and a novel vinylsulfone protease inhibitor) that can be used to treat patients suffering with the 2019-nCoV. We also summarize previous efforts into drugging these targets and hope to help in the development of broad spectrum anti-coronaviral agents for future epidemics.
Introduction
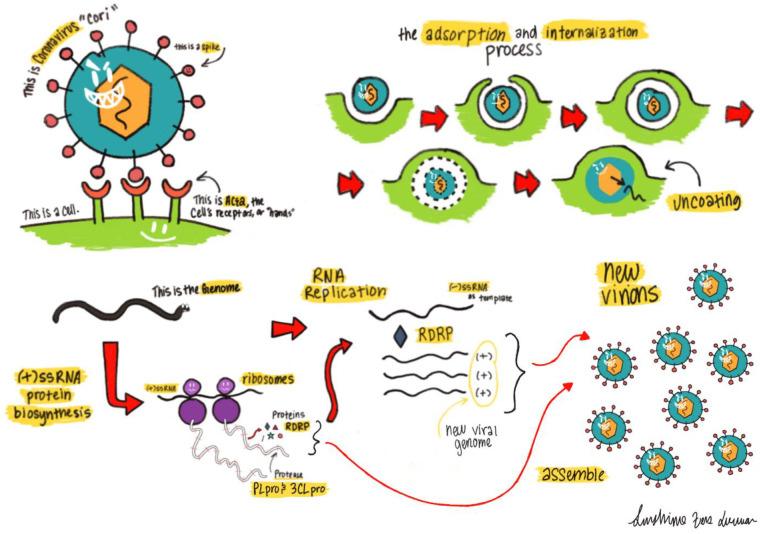
The 2019 novel coronavirus (2019-nCoV) is a newly emerged human-infectious coronavirus (CoV) that was originated in a Wuhan seafood market but has quickly spread in and beyond China.1 As of Jan 26th, 2019, there have been more than 2000 diagnosed cases and 56 confirmed deaths (Xinhua News). Since the pathogenesis of this virus is yet to be understood, there are scarce treatment options available to healthcare professionals who are fighting this epidemic at the front line. Praises need to be given to Chinese researchers who have acted quickly to isolate and sequence the virus. The availability of the virus genome sequence (GenBank ID: MN908947.3) makes it possible to identify treatments. Although it is essential to develop vaccines, small molecules, and biological therapeutics to specifically target the 2019-nCoV virus, it is unlikely that any effort made at the moment will benefit patients in the current outbreak. 2019-nCoV shares 82% sequence identity with severe acute respiratory syndrome-related coronavirus (SARS-CoV, GenBank ID: NC_004718.3) and more than 90% sequence identity in several essential enzymes (Figures 2–3, 5–6). What we have learned from several medicinal chemistry studies about SARS-CoV and the Middle East Resipatory Syndrome (MERS-CoV) may be directly used in helping us treat the 2019-nCoV. CoV relies on its spike protein to bind a host cell surface receptor for entry (Figure 1).2 For the 2019-nCoV, it is evident that this receptor is angiotensin-converting enzyme 2 (ACE2).3 After its entry into the host cell, the positive genomic RNA attaches directly to the host ribosome for translation of two large, co-terminal polyproteins that are processed by proteolysis to components for packing new virions.4 Two proteases that participate in this proteolysis process are the coronavirus main proteinase (3CLpro) and the papain-like protease (PLpro).5 In order to replicate the RNA genome, the CoV encodes a replicase that is an RNA-dependent RNA polymerase (RdRp).6 These four proteins are essential for the pathogen. Therapeutics currently targeting spike, RdRp, 3CLpro, and PLpro are possible treatments for 2019-nCoV. In this review, we will analyze similarities in spike, RdRp, 3CLpro, and PLpro proteins between the 2019-nCoV and SARS-CoV and suggest possible prevention and treatment options. Since little is known so far about the virulence of this virus, we will also discuss about the interactions between spike and ACE2 that may challenge the current view that 2019-nCoV is less virulent than SARS-CoV attributing to weaker interactions between spike and ACE2.
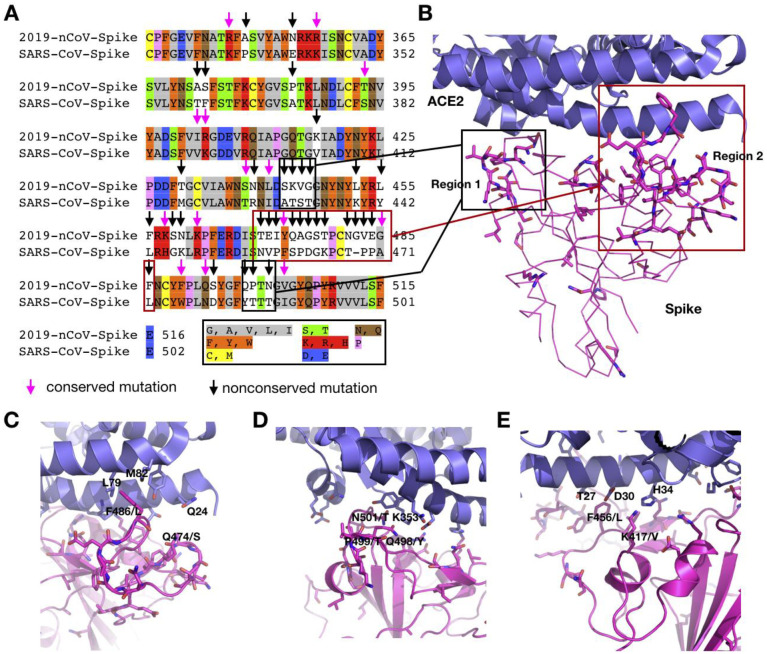
Figure 2.
A) Sequence alignment for the amino acids between the 2019-nCoV and SARS-CoV spike RBD domain. Conserved and non-conserved mutations are highlighted. B-E) Various binding interactions between the 2019-nCov spike protein (homology model built using Modeller, based upon PDB entry 2AJF) and ACE2 in regions 1 and 2
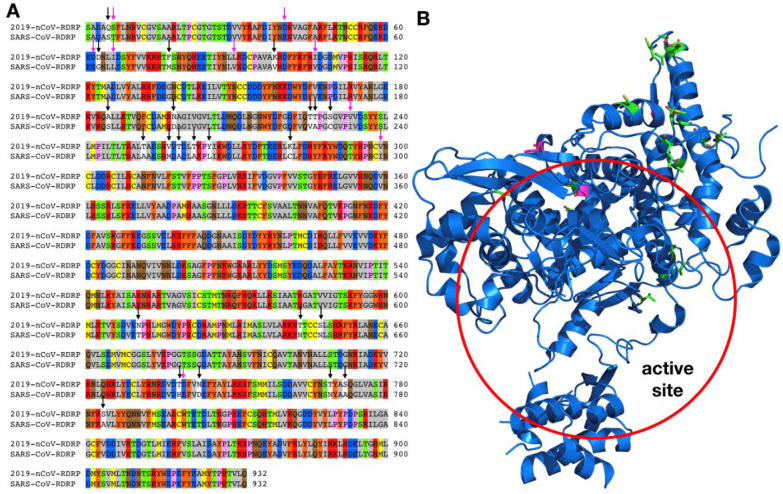
Figure 3.
A) Sequence alignment for the amino acids between the 2019-nCoV RdRp and the SARS-CoV RdRp. Conserved and non-conserved mutations are highlighted. B) Crystal structure of the SARS-CoV RdRp active site (PDB entry: 6NUS)
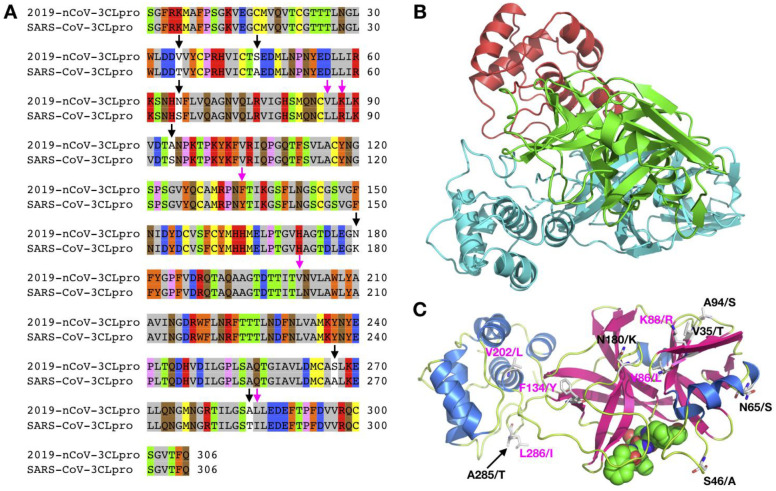
Figure 5.
A) Sequence alignment for the amino acids between the 2019-nCoV 3CLpro and the SARS-CoV 3CLpro. Conserved and non-conserved mutations are highlighted. B-C) A modeled 2019-nCoV 3CLpro structure using Modeller based on the SARS-CoV 3CLpro structure (PDB entry: 2A5I)
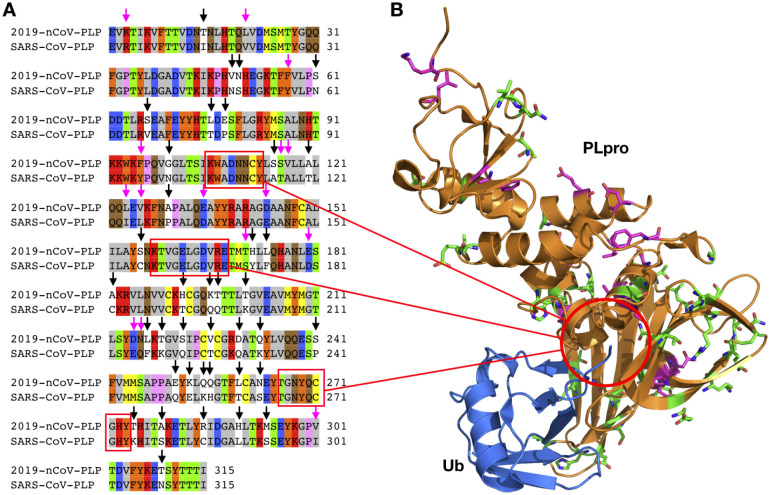
Figure 6.
A) Sequence alignment for the amino acids between the 2019-nCoV PLpro and the SARS-CoV PLpro. Conserved and non-conserved mutations are highlighted. B) Crystal structure of the SARS-CoV PLpro (PDB entry: 4MM3)
Figure 1.
Lifecycle of a coronavirus entering and replicating inside of a host cell. The (+)-stranded RNA is released upon viral entry, which starts the process of generating the viral coat and replicating the RNA genome
The Spike Protein
Both 2019-nCoV and SARS-CoV encode a large spike protein (2019-nCoV: 1253 aa; SARS-CoV: 1273 aa). The sequence identity of this protein between two origins is 76%. A large variation exists at the N-terminus (Figure 2A). The spike protein has three regions, S1, S2, and S3. For the SARS-CoV there is a receptor binding domain (RBD) in the S1 region that interacts with ACE2 with high affinity. The current assumption is that 2019-nCoV also engages this RBD to bind ACE2 for the entry into its human host cell. The alignment of the RBD from the two origins shows 73.5% sequence identity (Figure 2A). However, nonconserved mutations are highly accumulated in two structural regions (1 and 2) that interact directly with ACE2 (Figure 2B).2 Both crystal and cryo-EM structures of the SARS-CoV spike-ACE2 complex (PDB entries: 2AJF and 6ACD) have showed that only region 1 and region 2 engage in hydrogen bonding and hydrophobic interactions with ACE2. Since many residues in these two regions have been replaced in 2019-nCoV, this will lead to a loss in some of these interactions. It has also been predicted that the 2019-nCoV RBD interacts with ACE2 weaker than the SARS-CoV RBD.3 However, both regions are highly looped structures. Large variations in the two regions will inevitably lead to structural rearrangements that potentiate novel and possibly even stronger interactions with ACE2. For region 2, there is almost no similarity in the sequence between the two virus origins; however, it is premature to presume that the protein will fold in the same way to interact with ACE2. Assuming that region 2 of 2019-nCoV folds in the same way as in SARS-CoV, F486 will poise right in the position that engages strong hydrophobic interactions with both L79 and M82 in ACE2. These interactions do not exist in the SARS RBD-ACE2 complex due to the significant smaller L472 in that position. Another residue that potentially engages strong hydrogen bonding interactions is Q474 that is in the distance to engage a rearranged Q24 in ACE2. In region 1, a critical residue Y484 in SARS-CoV is replaced by Q498 in the 2019-nCoV. However, P499 that is a known secondary structure disruptor in the 2019-nCoV is expected to lead to a structural rearrangement at region 1 in the SARS-CoV RBD. In combination with Q498 and N501, new hydrogen bonding interactions that may involve K353 and other residues in ACE2 may form. Another notable difference between the 2019-nCoV and the SARS-CoV in RBD is K417(2019-nCoV)/V404(SARS-CoV). This is a residue in the middle of the concaved RBD binding surface that involves no interaction between SARS-CoV and ACE2. However, the long and positively charged potential of K417 makes it possible to engage strong hydrogen bonding and salt bridge interactions with H34 and D30 respectively in ACE2. Although molecular simulation may be used to analyze all these possible interactions in details, uncertainty will persist about them until the structure of the 2019-nCoV-RBD-ACE2 complex is determined using crystallography or cryo-EM. Given the urgency of the matter, there must be multiple research groups working on this. We hope to see their results within a short time. Alternatively, the 2019-nCoV RBD can be expressed and its affinity toward ACE2 independently determined biochemically and compared with that of the SARS-CoV RBD. Before any solid experimental results are available, any claims about weaker binding of the 2019-nCoV RBD toward ACE2 than that of the SARS-CoV RBD is premature.
The roles of the SARS-CoV spike protein in receptor binding and membrane fusion make it an ideal target for vaccine and antiviral development. The development of SARS vaccines based on the spike protein has been summarized in several previous reviews.7–11 Several strategies including live-attenuated SARS-CoV, killed SARS-CoV, DNA vaccines and viral vectored vaccines that have been successfully used to vaccinate against the animal SARS-CoVs.7, 12, 13 Similar ideas may be applied in developing 2019-nCoV vaccines. Alternative approaches are to directly use the 2019-nCoV RBD in combination with immunity-promoting adjuvants as a vaccine to trigger the human body to develop antibodies for the 2019-nCoV RBD neutralizing the virus.14
Although there are published results about therapeutic antibodies and peptides developed to neutralize the SARS-CoV spike protein, they are expected to have little use in neutralizing the 2019-nCoV. As discussed, above, the two engaging regions in the spike RBD for binding ACE2 are highly different between the SARS-CoV and 2019-nCoV. Antibodies and peptides targeting two regions in the SARS-CoV RBD are expected to interact weakly with the 2019-nCoV RBD. Novel antibodies and therapeutic peptides that interact potently with the 2019-nCoV RBD can be used to block its interaction with ACE2. Several research groups including ours have developed methods for building macrocyclic peptide libraries and apply them for the quick identification of macrocyclic peptide ligands for drug targets.15–20 Applications of these libraries to search for potent ligands for the 2019-nCoV RBD or the two ACE2 engaging peptide regions will potentially lead to rapid discovery of anti-2019-nCoV macrocyclic peptides. Although our group has initiated this effort, the lengthy process of the drug discovery process will make it not possible to help patients in the current epidemic. Learning from the study of SARS-CoV, a possible alternative is the direct use of peptides derived from the 2019-nCoV RBD and ACE2. Peptides derived from both the SARS-CoV RBD and ACE2 have been developed as novel therapeutics against the SARS-CoV infection by blocking the SARS-CoV RBD-ACE2 binding. For example, a peptide that overlaps the RBD sequence (aa 471–503) can specifically block the binding of ACE2 to the SARS-CoV RBD and inhibit the SARS-CoV entry into Vero cells with an EC50 of 41.6 μM.21 One peptide comprised of two ACE2 motifs (aa 22–44 and 351–357) linked by glycine, exhibited a potent anti-SARS activity with IC50 of 0.1 μM.22 Before any potent therapeutics to neutralize the 2019-nCoV RBD-ACE2 interaction are available, possible quick solutions to block this interaction is to use the 2019-nCoV RBD-based peptides and their combination cocktails.
RdRp
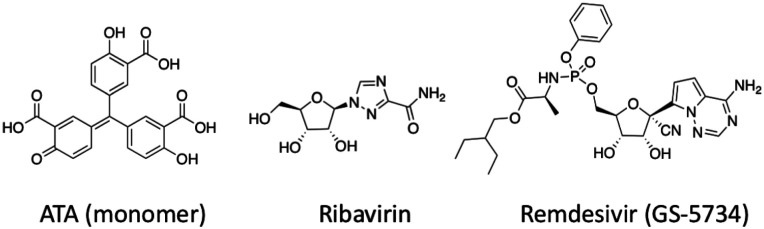
Although 2019-nCoV and SARS-CoV shares 82% sequence identity at their genomic RNA level, their RdRp proteins share remarkable 96% sequence identity (Figure 3A). RdRp involves a very large and deep groove as an active site for the polymerization of RNA. Residues that show variations between the 2019-nCoV and SARS-CoV RdRps are mostly distal to this active site (Figure 3B).23 This high sequence conservation between the two enzymes makes it highly likely that any potent agents developed for the SARS-CoV RdRp will exhibit equal potency and efficacy on the 2019-nCoV RdRp. Although not extensively explored, there do exist several agents that target the SARS-CoV RdRp or its catalyzed polymerization process. One such compound found to show anti-viral activity was aurintricarboxylic acid (ATA in Figure 4). ATA is an anionic polymer shown to bind to a variety of protein targets, including gp120 of HIV-1 and HIV-2, and was demonstrated to prevent SARS-CoV replication (EC50 = 0.2 mg / mL).24–26 Despite computational models validated against known ATA targets predicting RdRp as the bound target, no experimental evidence has demonstrated this relationship.27 Beyond this one exception, the remaining RdRp inhibitors have been nucleoside analogs, and these provide the most promising avenue towards disrupting viral RNA replication. The nucleoside analog ribavirin (RBV) has been tested against the SARS-CoV, and in SARS- and MERS-infected patients.28–31 At best, efficacy with RBV was inconclusive, with some studies showing a worsening of patient outcomes (as reviewed by Stockman, et al).32 Exonuclease activity by the enzyme nsp14 has been shown to be able to remove mismatches as well as incorporated nucleoside analogs, and inactivation of nsp14’s exonuclease activity has been shown to increase the efficacy of nucleosides like RBV.31, 33 In order for nucleoside analogs to effectively inhibit viral RNA replication, the nucleoside must either evade detection by the exonuclease or must outcompete exonuclease activity. Remdesivir (GS-5734) is an excellent example of the latter. An adenosine analog prodrug with a 1’-nitrile drug displayed potent efficacy against SARS and MERS in human airway epithelial (HAE) cell models and in mice (IC50 = 0.069 μM and 0.074 μM for SARS-CoV and MERS-CoV, respectively, in HAE).34 Activity against various bat coronaviruses was also demonstrated, with broad spectrum anti-coronavirus activity.34 Susceptibility of CoV to remdesivir was shown to be increased in strains with inactivated exonuclease activity.35 CoV resistance to remdesivir was studied in the model β-coronavirus, murine hepatitis virus (MHV). MHV passaged in the presence of the parent nucleoside, GS-441524 (containing a 6’-hydroxyl instead of a phosphoramidate), developed two mutations to the RdRp, F476L and V553L. These mutations conferred a 5.6-fold increase in resistance to remdesivir in MHV, and a 6-fold increase in resistance when the homologous mutations were introduced to the RdRp of SARS-CoV (0.01 μM vs 0.06 μM). Mice infected with this resistant SARS-CoV had significantly lower lung viral titers 4 days post-infection.35 Altogether, remdesivir has been shown to outcompete the proofreading ability of nsp12, and mutations which confer resistance attenuate virulence. Efforts towards drugging coronavirus RdRp in this manner should provide a basis not only to develop therapeutics for the 2019-nCoV but could provide for broad spectrum anti-virals useful for future CoV outbreaks.
Figure 4.
Structure of drugs inhibiting SARS-CoV viral replication via the mechanistic action of RdRp
3CLpro and PLpro
3CLpro and PLpro are two proteases that process the polypeptide translation from the genomic RNA to protein components either structural or non-structural for replication and packaging of new generation viruses. PLpro also serves as a deubiquitinase that function to deubiquitinate host cell proteins such as interferon factor 3 (IRF3) as well as to inactivate the pathway for nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB).36 This leads to immune suppression in the cells of the host being infected by the virus. Because both proteases are vital to the virus for replication and controlling the host cell, they are viable targets for antiviral agents. Similar to the RdRp protein, 2019-nCoV and SARS-CoV share remarkable 96% sequence identity in their decoded 3CLpro (Figure 5A). 3CLpro naturally forms a dimer and each monomer contains two regions, the N-terminal catalytic region and the C-terminal region (Figure 5B).36, 37 Most residues in the catalytic region that display variations between two origins are on the protein surface. Although S46(2019-nCoV)/A(SARS-CoV) may possibly interact with either substrates or inhibitors that bind to the active site, the small structural change from A to S is expected not to interfere significantly with the binding of small molecule inhibitors to the active site. Small molecule agents that potently inhibit the SARS-CoV 3CLpro are expected to function similarly toward the 2019-nCoV 3CLpro. Unlike 3CLpro, PLpro from the two origins shares only 83% sequence identity (Figure 6A). Residue variations between the two origins cover almost all over the surface of the PLpro. These substantial variations in amino acid compositions are expected to influence how the two PLpro enzymes interact with their ligands. However, the three secondary structure components that form the active site do not vary in the two PLpro proteins (Figure 6B).38 It is possible that an inhibitor developed for the SARS-CoV PLpro may also work for the 2019-nCoV PLpro.
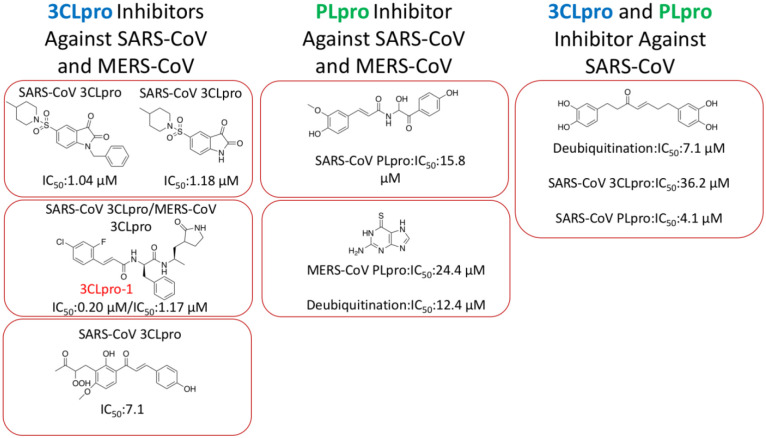
Over the last two decades, much of the research in drugging SAR-CoV has focused on the development of small molecule, peptide, and peptidomimetic inhibitors of 3CLpro and PLpro. Many of the inhibitors are in the μM range in terms of binding to and inhibiting the two proteases.39–44 However, there has been the identification of a few low nM range inhibitors that can be used in combination with other protease inhibitor therapies to help combat the virus.45 For the purpose of this section the classification of inhibitors will be divided into categories based on the proteases that are inhibited to stop the virus from taking control of the host cells. Each of the compounds was test in terms of either a SARS-CoV, MERS-CoV or deubiquitinate cell model. There have been several hundred small molecules developed to inhibit 3CLpro and PLpro, however these were the most potent since the early 2000s. The classifications with structures and inhibitory concentrations are summarized in Figure 7. These compounds are in the low μM range in terms of inhibition, leaving room for further development. However, there has already been extensive SAR performed on these final stage products guiding the researcher in knowing what substituents to modify when targeting the 2019-nCoV. This summary can also guide both researchers and health professionals in using combinational therapy with two or more of these compounds, as this has already been done in terms of treating people suffering with a CoV infection. One of these compounds (highlighted as 3CLpro-1 in Figure 7) has an IC50 value against the SARS-CoV of 200 nM.41 This potency may be adequate to combat 2019-nCoV.
Figure 7.
This is a representation of the top CoV protease inhibitors providing a scaffold to perform SAR in terms of design novel small molecule protease inhibitors for 2019-nCoV40,41,42,43,47,39,44. 3CLpro-1 is highlight as the most potent inhibitor
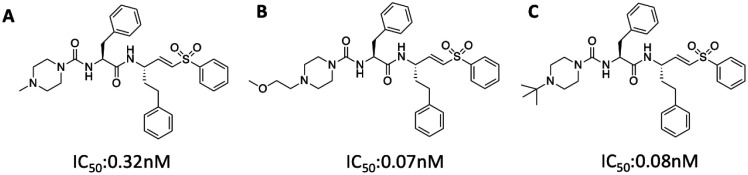
Both 3CLpro and PLpro are cysteine proteases. Covalent inhibitors with high potencies can be potentially developed for them. Recently Zhou and coworkers develop a class of potentially covalent cysteine protease inhibitors that specifically target the coronavirus entry.45 There was no direct relationship to 3CLpro and PLpro, however this class of vinylsulfone small molecules was able to inhibit replication of the virus in the nM range. This group discovered that inhibition of serine proteases (using camostat) in combination with cysteine protease (using their vinylsulfone protease inhibitors) is able to combat the SARS-CoV. The survival of mice suffering with SARS-CoV significantly increased in comparison to the control group when treated with this combination therapy. They studied several variations of vinylsulfone small molecules which are shown below with their corresponding IC50 values In Figure 8 (compound A to C). Once again, these vinylsulfone small molecules provide an additional scaffold for SAR development. They can also be tested against the specific 3CLpro and PLpro in order to further elucidate their specific mechanism of action. Given their high potency against SARS-CoV, it is possible that they are equally potent against the 2019-nCoV.
Figure 8.
Lead vinylsulfone protease inhibitors that prevent the entry of the CoV and in combination with camostat increase the survival rate of a mice model suffering with SARS-CoV infection
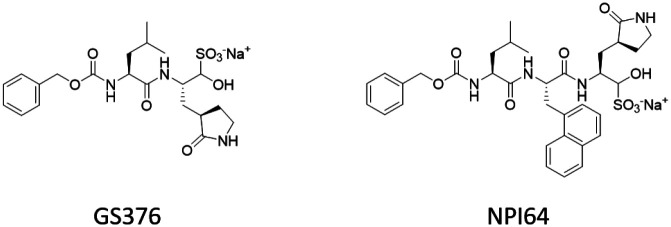
Looking to more distant members of the orthocoronavinae subfamily can also provide inspiration towards new therapeutic regimens, such as with studies done with feline coronavirus (FCoV) and its mutated form, feline infectious peritonitis virus (FIPV). The tripeptidyl bisulfite adducts GC376 and NPI64 (Figure 9) were found to be potent inhibitors of FIPV replication at 0.04 μM for both drugs.46 3CLpro of both FIPV and SARS-CoV share about 50% sequence identity, but the overall structure is conserved; in a FRET-based activity assay, the IC50 value of GC376 against 3CLpro was found to be 4.9-fold in SARS-CoV compared to FIPV. In combination with a high compound cytotoxicity (CC50 > 150 μM and CC50 = 61.91 μM in CRFK cells for GC376 and NPI64, respectively), these masked-aldehyde warheads should be investigated for efficacy on the 3CLpro of 2019-nCoV as soon as possible.
Figure 9.
Peptidyl bisulfide adducts which have been demonstrated to prevent viral replication in the feline coronavirus FIPV. GC376 (left) was shown to produce similar levels of inhibition against SARS-CoV 3CLpro in a FRET-based activity assay
Conclusions
The 2019-nCoV and SARS-CoV share very high sequence identity on their RdRp and 3CLpro proteins. Previous efforts have resulted in the discovery of some potent small molecule therapeutics based on these two proteins in the SARS-CoV. We envision that remdesivir and 3CLpro-1 may be directly applied to treat the 2019-nCoV. Since remdesivir is a drug undergoing a clinical trial, the authority in China may negotiate with Gilead in possible use of this drug for patients suffering with the 2019-nCoV. Other potential small molecule therapeutics for the 2019-nCoV are the molecules shown in Figures 8 and 9. The 2019-nCoV spike RBD is significantly different from the SARS-CoV spike RBD especially in two regions when binding to ACE2. This difference effectively rules out the use of previously developed antibodies and therapeutic peptides for the SARS-CoV spike RBD. However, a possible quick solution to inhibit the RBD-ACE2 interaction for preventing the infection is to use peptides and their cocktails derived from RBD and ACE2.
Acknowledgement
Research support in the Texas A&M Drug Discovery Center is provided from National Institute of Health (grants R01GM121584 and R01GM127575), Cancer Prevention & Research Institute of Texas (grant RP170797), and Welch Foundation (grant A-1715). We thank Sunshine Zea Leeuwon for providing graphical assistance in designing all the figures.
References
- 1.Zhou P.; Yang X. L.; Wang X. G.; Hu B.; Zhang L.; Zhang W.; Si H. R.; Zhu Y.; Li B.; Huang C. L.; Chen H. D.; Chen J.; Luo Y.; Guo H.; Jiang R. D.; Liu M. Q.; Chen Y.; Shen X. R.; Wang X.; Zheng X. S.; Zhao K.; Chen Q. J.; Deng F.; Liu L. L.; Yan B.; Zhan F. X.; Wang Y. Y.; Xiao G.; Shi Z. L. Discovery of a Novel Coronavirus Associated with the Recent Pneumonia Outbreak in Humans and its Potential Bat Origin. 2020. bioRxiv 2020.01.22.914952. https://www.biorxiv.org/content/10.1101/2020.01.22.914952v2 (accessed Jan 25, 2020). [Google Scholar]
- 2.Li F.; Li W.; Farzan M.; Harrison S. C. Structure of SARS Coronavirus Spike Receptor-Binding Domain Complexed with Receptor. Science 2005, 309, 1864–1868. [DOI] [PubMed] [Google Scholar]
- 3.Dong N.; Yang X.; Ye L.; Chen K.; Chan E. W. C.; Yang M.; Chen S. Genomic and Protein Structure Modelling Analysis Depicts the Origin and Infectivity of 2019-nCoV, a New Coronavirus which Caused a Pneumonia Outbreak in Wuhan, China. 2020. bioRxiv 2020.01.20.913368. https://www.biorxiv.org/content/10.1101/2020.01.20.913368v2 (accessed Jan 25, 2020). [Google Scholar]
- 4.Baranov P. V.; Henderson C. M.; Anderson C. B.; Gesteland R. F.; Atkins J. F.; Howard M. T. Programmed Ribosomal Frameshifting in Decoding the SARS-CoV Genome. Virology 2005, 332, 498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ziebuhr J.; Snijder E. J.; Gorbalenya A. E. Virus-Encoded Proteinases and Proteolytic Processing in the Nidovirales. J. Gen. Virol. 2000, 81, 853–879. [DOI] [PubMed] [Google Scholar]
- 6.Xu X.; Liu Y.; Weiss S.; Arnold E.; Sarafianos S. G.; Ding J. Molecular Model of SARS Coronavirus Polymerase: Implications for Biochemical Functions and Drug Design. Nucleic Acids Res 2003, 31, 7117–7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavanagh D. Severe Acute Respiratory Syndrome Vaccine Development: Experiences of Vaccination Against Avian Infectious Bronchitis Coronavirus. Avian Pathol. 2003, 32, 567–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du L.; He Y.; Zhou Y.; Liu S.; Zheng B. J.; Jiang S. The Spike Protein of SARS-CoV--a Target for Vaccine and Therapeutic Development. Nat. Rev. Microbiol. 2009, 7, 226–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang S.; He Y.; Liu S. SARS Vaccine Development. Emerg. Infect. Dis. 2005, 11, 1016–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor D. R. Obstacles and Advances in SARS Vaccine Development. Vaccine 2006, 24, 863–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roper R. L.; Rehm K. E. SARS Vaccines: Where Are We? Expert. Rev. Vaccines. 2009, 8, 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes K. V. SARS Coronavirus: a New Challenge for Prevention and Therapy. J. Clin. Invest. 2003, 111, 1605–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navas-Martin S. R.; Weiss S. Coronavirus Replication and Pathogenesis: Implications for the Recent Outbreak of Severe Acute Respiratory Syndrome (SARS), and the Challenge for Vaccine Development. J. Neurovirol. 2004, 10, 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Y.; Zhou Y.; Liu S.; Kou Z.; Li W.; Farzan M.; Jiang S. Receptor-Binding Domain of SARS-CoV Spike Protein Induces Highly Potent Neutralizing Antibodies: Implication for Developing Subunit Vaccine. Biochem. Biophys. Res. Commun. 2004, 324, 773–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Neil K. T.; Hoess R. H.; Jackson S. A.; Ramachandran N. S.; Mousa S. A.; DeGrado W. F. Identification of Novel Peptide Antagonists for GPIIb/IIIa from a Conformationally Constrained Phage Peptide Library. Proteins 1992, 14, 509–515. [DOI] [PubMed] [Google Scholar]
- 16.McLafferty M. A.; Kent R. B.; Ladner R. C.; Markland W. M13 Bacteriophage Displaying Disulfide-Constrained Microproteins. Gene 1993, 128, 29–36. [DOI] [PubMed] [Google Scholar]
- 17.Hipolito C. J.; Suga H. Ribosomal Production and In Vitro Selection of Natural Product-Like Peptidomimetics: The FIT and RaPID Systems. Curr. Opin. in Chem. Biol. 2012, 16, 196–203. [DOI] [PubMed] [Google Scholar]
- 18.Palei S.; Becher K. S.; Nienberg C.; Jose J.; Mootz H. D. Bacterial Cell-Surface Display of Semisynthetic Cyclic Peptides. ChemBioChem 2019, 20, 72–77. [DOI] [PubMed] [Google Scholar]
- 19.Xiao W.; Wang Y.; Lau E. Y.; Luo J.; Yao N.; Shi C.; Meza L.; Tseng H.; Maeda Y.; Kumaresan P.; Liu R.; Lightstone F. C.; Takada Y.; Lam K. S. The Use of One-Bead One-Compound Combinatorial Library Technology to Discover High-Affinity αvβ3 Integrin and Cancer Targeting Arginine-Glycine-Aspartic Acid Ligands with a Built-in Handle. Mol. Cancer Ther. 2010, 9, 2714–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X. S.; Chen P. H. C.; Hampton J. T.; Tharp J. M.; Reed C. A.; Das S. K.; Wang D. S.; Hayatshahi H. S.; Shen Y.; Liu J.; Liu W. R. A Genetically Encoded, Phage-Displayed Cyclic-Peptide Library. Angew. Chem. Int. Ed. 2019, 58, 15904–15909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu H.; Li L.; Kao R. Y.; Kou B.; Wang Z.; Zhang L.; Zhang H.; Hao Z.; Tsui W. H.; Ni A.; Cui L.; Fan B.; Guo F.; Rao S.; Jiang C.; Li Q.; Sun M.; He W.; Liu G. Screening and Identification of Linear B-Cell Epitopes and Entry-Blocking Peptide of Severe Acute Respiratory Syndrome (SARS)-Associated Coronavirus Using Synthetic Overlapping Peptide Library. J. Comb. Chem. 2005, 7, 648–656. [DOI] [PubMed] [Google Scholar]
- 22.Han D. P.; Penn-Nicholson A.; Cho M. W. Identification of Critical Determinants on ACE2 for SARS-CoV Entry and Development of a Potent Entry Inhibitor. Virology 2006, 350, 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirchdoerfer R. N.; Ward A. B. Structure of the SARS-CoV Nsp12 Polymerase Bound to Nsp7 and Nsp8 Co-Factors. Nat. Commun. 2019, 10, 2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.González R. G.; Blackburn B. J.; Schleich T. Fractionation and Structural Elucidation of the Active Components of Aurintricarboxylic Acid, a Potent Inhibitor of Protein Nucleic Acid Interactions. Biochim. Biophys. Acta, Nucleic Acids Protein Synth. 1979, 562, 534–545. [DOI] [PubMed] [Google Scholar]
- 25.Cushman M.; Wang P.; Chang S. H.; Wild C.; De Clercq E.; Schols D.; Goldman M. E.; Bowen J. A. Preparation and Anti-HIV Activities of Aurintricarboxylic Acid Fractions and Analogs: Direct Correlation of Antiviral Potency with Molecular Weight. J. Med. Chem. 1991, 34, 329–337. [DOI] [PubMed] [Google Scholar]
- 26.He R.; Adonov A.; Traykova-Adonova M.; Cao J.; Cutts T.; Grudesky E.; Deschambaul Y.; Berry J.; Drebot M.; Li X. Potent and Selective Inhibition of SARS Coronavirus Replication by Aurintricarboxylic Acid. Biochem. Biophys. Res. Commun. 2004, 320, 1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yap Y.; Zhang X.; Andonov A.; He R. Structural Analysis of Inhibition Mechanisms of Aurintricarboxylic Acid on SARS-CoV Polymerase and Other Proteins. Comput. Biol. Chem. 2005, 29, 212–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiou H. E.; Liu C. L.; Buttrey M. J.; Kuo H. P.; Liu H. W.; Kuo H. T.; Lu Y. T. Adverse Effects of Ribavirin and Outcome in Severe Acute Respiratory Syndrome: Experience in Two Medical Centers. Chest 2005, 128, 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muller M. P.; Dresser L.; Raboud J.; McGeer A.; Rea E.; Richardson S. E.; Mazzulli T.; Loeb M.; Louie M.; Canadian S. R. N. Adverse Events Associated with High-Dose Ribavirin: Evidence from the Toronto Outbreak of Severe Acute Respiratory Syndrome. Pharmacotherapy 2007, 27, 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Tawfiq J. A.; Momattin H.; Dib J.; Memish Z. A., Ribavirin and Interferon Therapy in Patients Infected with the Middle East Respiratory Syndrome Coronavirus: an Observational Study. J. Infect. Dis. 2014, 20, 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith E. C.; Blanc H.; Vignuzzi M.; Denison M. R. Coronaviruses Lacking Exoribonuclease Activity Are Susceptible to Lethal Mutagenesis: Evidence for Proofreading and Potential Therapeutics. PLoS Pathog. 2013, 9, e1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stockman L. J.; Bellamy R.; Garner P. SARS: Systematic Review of Treatment Effects. PLoS Med. 2006, 3, e343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferron F.; Subissi L.; De Morais A. T. S.; Le N. T. T.; Sevajol M.; Gluais L.; Decroly E.; Vonrhein C.; Bricogne G.; Canard B.; Imbert I. Structural and Molecular Basis of Mismatch Correction and Ribavirin Excision from Coronavirus RNA. P. Natl. Acad. Sci. USA 2018, 115, E162–E171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheahan T. P.; Sims A. C.; Graham R. L.; Menachery V. D.; Gralinski L. E.; Case J. B.; Leist S. R.; Pyrc K.; Feng J. Y.; Trantcheva I.; Bannister R.; Park Y.; Babusis D.; Clarke M. O.; Mackman R. L.; Spahn J. E.; Palmiotti C. A.; Siegel D.; Ray A. S.; Cihlar T.; Jordan R.; Denison M. R.; Baric R. S. Broad-Spectrum Antiviral GS-5734 Inhibits Both Epidemic and Zoonotic Coronaviruses. Sci. Transl. Med. 2017, 9, eaal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agostini M. L.; Andres E. L.; Sims A. C.; Graham R. L.; Sheahan T. P.; Lu X.; Smith E. C.; Case J. B.; Feng J. Y.; Jordan R.; Ray A. S.; Cihlar T.; Siegel D.; Mackman R. L.; Clarke M. O.; Baric R. S.; Denison M. R. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio 2018, 9, e00221–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Báez-Santos Y. M.; St. John S. E.; Mesecar A. D. The SARS-Coronavirus Papain-Like Protease: Structure, Function and Inhibition by Designed Antiviral Compounds. Antivir. Res. 2015, 115, 21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee T. W.; Cherney M. M.; Huitema C.; Liu J.; James K. E.; Powers J. C.; Eltis L. D.; James M. N. Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-Like Aza-Peptide Epoxide. J. Mol. Biol. 2005, 353, 1137–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ratia K.; Kilianski A.; Baez-Santos Y. M.; Baker S. C.; Mesecar A. Structural Basis for the Ubiquitin-Linkage Specificity and deISGylating activity of SARS-CoV papain-like protease. PLoS Pathog. 2014, 10, e1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X.; Chou C. Y.; Chang G. G. Thiopurine Analogue Inhibitors of Severe Acute Respiratory Syndrome-Coronavirus Papain-Like Protease, a Deubiquitinating and deISGylating Enzyme. Antiviral Chem. Chemother. 2009, 19, 151–156. [DOI] [PubMed] [Google Scholar]
- 40.Cheng K. W.; Cheng S. C.; Chen W. Y.; Lin M. H.; Chuang S. J.; Cheng I. H.; Sun C. Y.; Chou C. Y., Thiopurine Analogs and Mycophenolic Acid Synergistically Inhibit the Papain-Like Protease of Middle East Respiratory Syndrome Coronavirus. Antivir. Res. 2015, 115, 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar V.; Shin J. S.; Shie J. J.; Ku K. B.; Kim C.; Go Y. Y.; Huang K. F.; Kim M.; Liang P. H. Identification and Evaluation of Potent Middle East Respiratory Syndrome Coronavirus (MERS-CoV) 3CLPro Inhibitors. Antivir. Res. 2017, 141, 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu W.; Zhu H. M.; Niu G. J.; Shi E. Z.; Chen J.; Sun B.; Chen W. Q.; Zhou H. G.; Yang C. Synthesis, Modification and Docking Studies of 5-Sulfonyl Isatin Derivatives as SARS-CoV 3C-Like Protease Inhibitors. Bioorg. Med. Chem. 2014, 22, 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park J. Y.; Jeong H. J.; Kim J. H.; Kim Y. M.; Park S. J.; Kim D.; Park K. H.; Lee W. S.; Ryu Y. B. Diarylheptanoids from Alnus japonica Inhibit Papain-Like Protease of Severe Acute Respiratory Syndrome Coronavirus. Biol. Pharm. Bull. 2012, 35, 2036–2042. [DOI] [PubMed] [Google Scholar]
- 44.Park J. Y.; Ko J. A.; Kim D. W.; Kim Y. M.; Kwon H. J.; Jeong H. J.; Kim C. Y.; Park K. H.; Lee W. S.; Ryu Y. B. Chalcones Isolated from Angelica Keiskei Inhibit Cysteine Proteases of SARS-CoV. J. Enzym. Inhib. Med. Chem. 2016, 31, 23–30. [DOI] [PubMed] [Google Scholar]
- 45.Zhou Y.; Vedantham P.; Lu K.; Agudelo J.; Carrion R. Jr.; Nunneley J. W.; Barnard D.; Pohlmann S.; McKerrow J. H.; Renslo A. R.; Simmons G. Protease Inhibitors Targeting Coronavirus and Filovirus Entry. Antivir. Res. 2015, 116, 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim Y.; Shivanna V.; Narayanan S.; Prior A. M.; Weerasekara S.; Hua D. H.; Kankanamalage A. C. G.; Groutas W. C.; Chang K. O. Broad-Spectrum Inhibitors against 3C-Like Proteases of Feline Coronaviruses and Feline Caliciviruses. J. Virol. 2015, 89, 4942–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song Y. H.; Kim D. W.; Curtis-Long M. J.; Yuk H. J.; Wang Y.; Zhuang N.; Lee K. H.; Jeon K. S.; Park K. H. Papain-Like Protease (PLpro) Inhibitory Effects of Cinnamic Amides from Tribulus terrestrisFruits. Biol. Pharm. Bull. 2014, 37, 1021–1028. [DOI] [PubMed] [Google Scholar]