Figure 1.
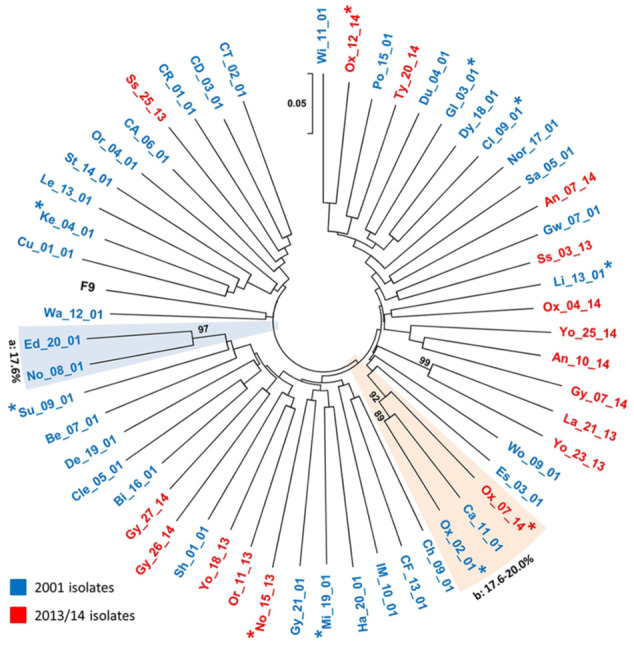
Unrooted neighbour-joining tree of the 56 partial feline calicivirus (FCV) capsid sequences used in this study (including FCV-F9; GenBank Accession Number M86379). The evolutionary distances were computed using the p-distance method, 20 and are in the units of the number of base differences per site (see 0.05 scale bar, which equates to five changes per 100 bases). All codon positions were included. There were 432 nucleotide positions in the final data set. The isolates are numbered as in Table 1 in the supplementary material, including a two-letter code for the geographical area, two digits for the isolate number and two digits for the year of collection; isolates from each of the two studies are also differentiated by colour (see key).
Asterisks represent isolates from cats with acute disease (upper respiratory tract disease ± ulcers). Clades represented by more than a single sequence (<20% capsid divergence, ⩾80% bootstrap values) are boxed, additionally labelled a and b, and the intra-clade diversity indicated. Bootstrap values <80% are not shown
