Abstract
In this issue of Structure, Coulombe and coworkers (Lee et al., 2020) present the crystal structure of the keratin 5/14 2B heterodimeric complex containing the keratin 14 substitution C367A. The authors identify a 2B-2B contact interface important to the elongation of mature keratin 5/14 filaments.
Although keratins were one of the first proteins to be analyzed by x-ray diffraction by W.T. Astbury and A. Street in 1932 (University of Leeds, UK), our current understanding of keratin intermediate filament (KIF) structural biology lags significantly behind that of other cytoskeletal elements. Almost 90 years after Astbury’s studies, there are only six human keratin x-ray crystal structures deposited in the Protein Data Bank (PDB) and they account for only 4 out of 54 distinct keratins expressed in humans. Intermediate filaments (IFs) have been identified as therapeutic targets for the treatment of malignancies and other human diseases; however, no IF-targeting drugs are currently used in routine clinical practice. This is in contrast to microtubule-targeting drugs which have been established as standard chemotherapeutic drugs for many years. Furthermore, mutations in IFs are responsible for multiple human diseases, but the overall lack of mechanistic insight into IF function hinders the development of effective treatments.
One barrier to developing clinically viable IF-specific drugs is our limited understanding of how IFs assemble at the molecular level. Our current knowledge of IF assembly is rooted in chemical cross- linking and nearest neighbor analysis, which identified four modes (A11, A22, A12, and ACN) in which IF dimers align into tetramers (Geisler et al., 1992; Steinert et al., 1993). Subsequent small-angle x-ray scattering (SAXS) studies identified the formation of octamer and 32mer units in the process of IF assembly (Sokolova et al., 2006). X-ray crystallography has played a key role in understanding the molecular mechanisms behind these observations, as crystal structures have provided insight into the A11 and ACN modes, but there has been a significant lack of structural information addressing the molecular contacts that drive the A12 or A22 modes of interaction.
In the context of keratin 5-keratin 14 (K5/K14) biology, Coulombe and colleagues previously determined a wild-type 2B domain structure that resulted in a unique finding at cysteine 367 of K14 (Lee et al., 2012). An interdimer disulfide bond occurred between K14 molecules from two K5/K14 heterodimers creating an “X-shaped” complex. This structure revealed a biological function for disulfide bonding in forming a “cage” of IFs around the nucleus (Lee et al., 2012). In a follow-up study, they demonstrated that interkeratin disulfide bonding regulates perinuclear KIF networks, as well as cellular dynamics and cycling of keratins (Feng and Coulombe, 2015). Building from this work, in the current issue of Structure Coulombe and colleagues mutate K14-Cys367 to alanine (to alter the 2B crystal lattice) and determine the crystal structure of the K5/K14C367A 2B complex (PDB ID: 6JFV). They used the molecular contacts in this new crystal lattice to identify 3 key inter-dimer (ID) interfaces and hypothesize that they might drive in vivo filament assembly. Using cell imaging and negative stain electron microscopy (EM), they determine that mutations in one of these ID interfaces impairs the ability of keratin filaments to elongate; this interface is termed “ID1” and occurs between the N- and C- termini of two adjacent 2B heterodimers. These findings are supported by a crystal structure of the keratin 1- keratin 10C401A 2B domain, which identified that the C-terminus of the 2B domain is a hotspot for intermolecular interactions (Lomakin et al., 2020) (PDB ID: 6UUI). Some residues in the C-terminus of the 2B domain are highly conserved across IFs, which underscores its potential role in intermolecular interactions.
These findings are consistent with an assembly model based primarily on the cross-linking studies by Steinert and colleagues in which the 2B interactions in the A22 alignment may stabilize the octameric or higher associations in the IF (Steinert et al, 1993). This model is shown in the context of our current understanding of IF production, assembly, and dynamics in the cell (Figure 1). Several pathologic K5 and K14 mutations exist in the ID1 interface and are known to cause the human skin disease epidermolysis bullosa simplex (EBS), therefore increasing the potential importance and clinical relevance of the ID1 interface. EBS is characterized by the formation of blisters due to fracturing of basal keratinocytes. In healthy keratinocytes, K5/K14 filaments anchor the cell to the extracellular matrix via hemidesmosome interactions. Disruption of K5/K14 filaments in EBS causes loss of cell integrity between the nucleus and the hemidesmosome. Analysis of K5/K14 filaments containing K14 mutations associated with the severe generalized subtype of EBS (e.g. EBS-Dowling-Meara) demonstrated shorter filament lengths in vitro and K5/K14 aggregates in patient cells (Coulombe et al., 1991). These previous findings are in agreement with the current imaging results from EM and NIH 3T3 cell-based experiments on ID1 mutant filaments. Given that several of the known mutations in the ID1 interface cause severe generalized EBS, it is likely that K5 and K14 mutations in this region cause EBS by disrupting the elongation of K5/K14 filaments. The resulting short filaments are prone to aggregation and result in fragile keratinocytes because they do not provide sufficient mechanical strength. This work by Lee et al., (2020) serves as an example of how structural biology can help explain the link between genotype and phenotype–a concept we call “genotype-structurotype-phenotype” correlation (Figure 2) (Lomakin et al., 2020). This model for understanding the relationship between genetics, protein structure, and clinical presentation is widely applicable to the IF field because mutations in IFs cause over 80 human diseases.
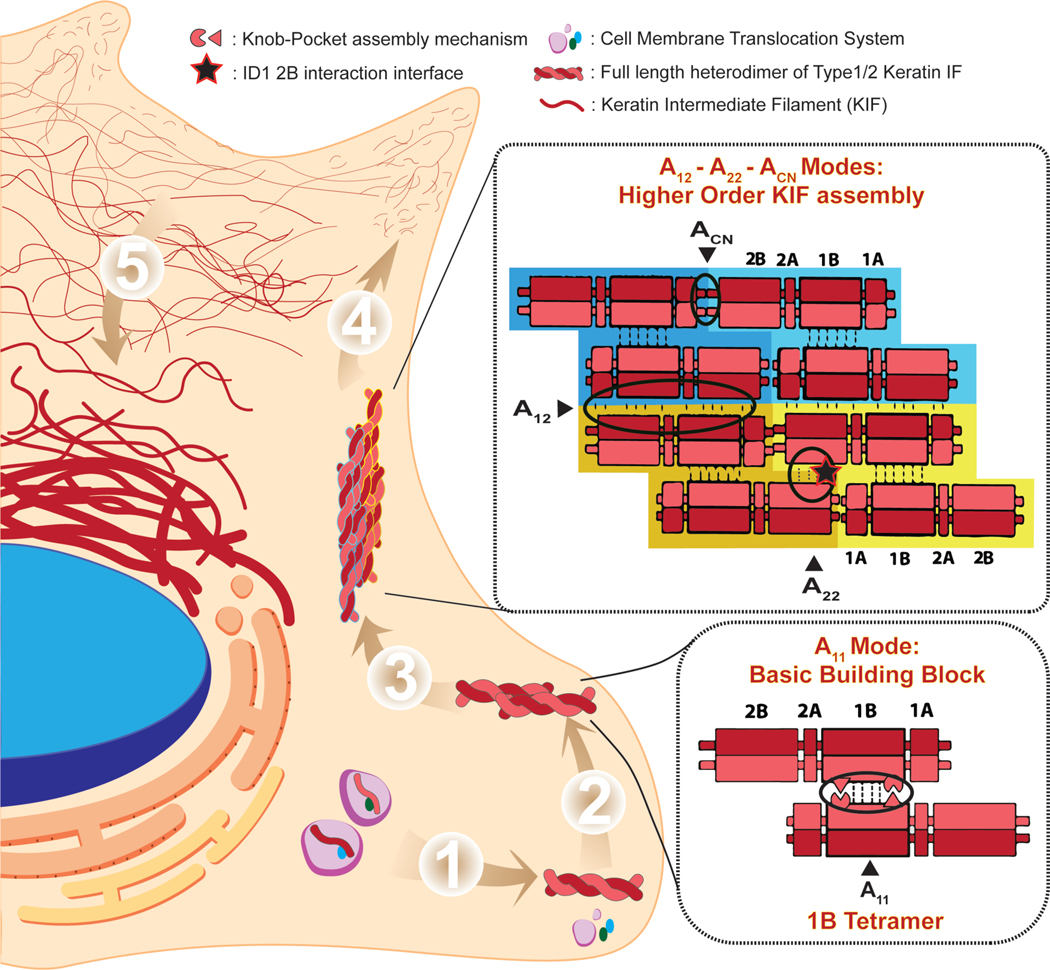
Figure 1. Keratin Intermediate Filament Assembly and Cellular Dynamics.
1) After being translated, the individual keratin monomer (IF Type 1/2) is bound to corresponding membrane translocation proteins to be moved to the cell periphery. Upon arrival at the cell membrane, the disassociation of membrane translocation proteins leads to obligate heterodimer formation between Type 1 and Type 2 keratins in parallel coiled-coil fashion. 2) Heterodimers then come together to form an anti-parallel tetramer, the basic building block of mature IFs, which is largely driven and stabilized by a knob-pocket assembly mechanism in the 1B helical domain (Eldirany et al., 2019). 3) The tetramers further assemble to form octamers longitudinally through modes of interaction designated as ACN and A22. Two octamers join laterally in anti-parallel fashion through a mode of interaction designated as A12 (Steinert et al., 1993). 4) Further association of octameric units leads to formation of a protofibril/Unit Length Filament (ULF). Several ULFs oligomerize or nucleate into small “particles,” which then assemble together to elongate into short filaments (also called “squiggles”). The short filaments move toward the cell center by an actin-dependent transport (Robert et al., 2016). Final maturation of the IF occurs with significant elongation followed by radial compaction to obtain the 10-nm diameter filament in the peripheral cytoplasm. Mature IFs incorporate into the peripheral keratin network. At this stage subunit exchange along the IF can occur to optimize the structural integrity needs of the cell. 5) As the KIFs continue to migrate inward toward the cell nucleus, they bundle together to create a tight KIF matrix around the cell nucleus.
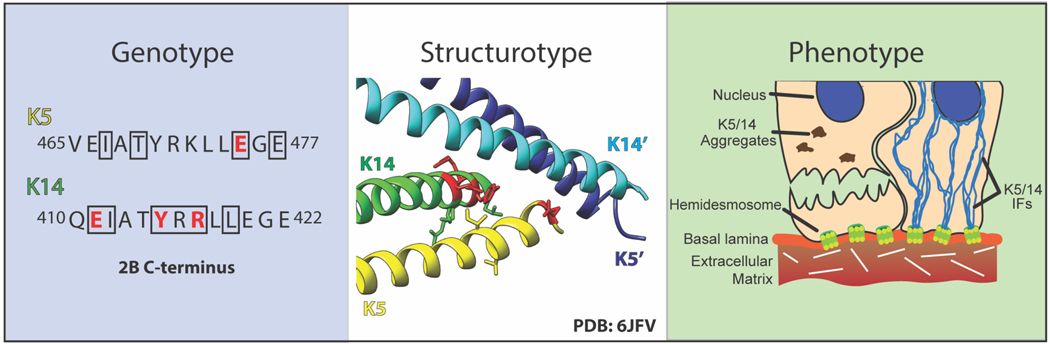
Figure 2. Genotype-Structurotype-Phenotype Correlation Applied to EBS-DM.
In the genotype panel, the protein sequence of the C-terminus of the K5/K14 2B domain is shown. The amino acid sequence represents the transcribed and translated “genotype” from the underlying DNA template from a patient. Enclosed in a black box are the residues for which missense mutations are known to cause severe generalized EBS (EBS-DM). Of those residues, the ones involved in the ID1 interface are colored red. No mutations causing EBS-DM have been identified in the N-terminus of the 2B domain. The structurotype panel shows the 2B C-terminus of a K5/14 dimer interacting with the N-terminus of a second dimer (K5’/14’) as observed in the high resolution crystal structure (PDB ID: 6JFV) by Lee et al. Residues causing EBS-DM are shown in stick rendering and those that participate in the ID1 interface are colored red. The EBS-DM phenotype is characterized histologically by K5/K14 aggregates and fractured cells that result medically in skin fragility and blistering in human patients. The phenotype illustration shows a fractured basal keratinocyte alongside a cell with a normal K5/K14 filament network for comparison.
Coulombe and colleagues provide a 2B structure and analysis that furthers our understanding of higher-order IF assembly by delineating critical 2B-2B interactions within a filament (Lee et al., 2020). Their work highlights the need for additional experimentally determined IF structures to fully elucidate IF assembly mechanisms. We believe that continued advancement in our understanding of the structural mechanisms of IF assembly can pave the way to IF-specific therapies. Identification of key interfaces like ID1 in the 2B domain (Lee et al., 2020) or the knob-pocket tetramerization mechanism in the 1B domain (Eldirany et al., 2019) present new opportunities for thinking about IF-based therapeutics. However, as the expansion of the number of experimentally determined high-resolution IF structures continues, it is important to avoid over-interpreting crystal lattice contacts and their relevance to IF biology. Crystal lattices often contain artifactual contacts not applicable to the real-life behavior of molecules; therefore, utilizing solution biophysics approaches, filament assembly assays, and cell biological studies are important ways to validate crystallographic findings so that the correct conclusions about biological relevance can be made. A strength of the article by Coulombe and colleagues (Lee et al., 2020) is their use of other scientific techniques (EM, cell-based filament network imaging) to complement their structural analysis. With this rigorous scientific approach, knowledge from the keratin “structurotype” will increase our understanding of how genetic alterations in patients lead to phenotypic disease.
Acknowledgements
This work was supported by the Richard K. Gershon, M.D., Student Research Fellowship at Yale University School of Medicine (to SAE) and an NIH/NIAMS Award K08-AR070290 (to CGB).
References
- Coulombe PA, Hutton ME, Letai A, Hebert A, Paller AS, and Fuchs E (1991). Point mutations in human keratin 14 genes of epidermolysis bullosa simplex patients: genetic and functional analyses. Cell 66, 1301–1311. [DOI] [PubMed] [Google Scholar]
- Eldirany SA, Ho M, Hinbest AJ, Lomakin IB, and Bunick CG (2019). Human keratin 1/10–1B tetramer structures reveal a knob-pocket mechanism in intermediate filament assembly. EMBO J 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, and Coulombe PA (2015). A role for disulfide bonding in keratin intermediate filament organization and dynamics in skin keratinocytes. J Cell Biol 209, 59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler N, Schünemann J, and Weber K (1992). Chemical cross-linking indicates a staggered and antiparallel protofilament of desmin intermediate filaments and characterizes one higher-level complex between protofilaments. Eur J Biochem 206, 841–852. [DOI] [PubMed] [Google Scholar]
- Lee CH, Kim MS, Chung BM, Leahy DJ, and Coulombe PA (2012). Structural basis for heteromeric assembly and perinuclear organization of keratin filaments. Nat Struct Mol Biol 19, 707–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Kim MS, Li S, Leahy DJ, and Coulombe PA (2020). Structure-Function Analyses of a Keratin Heterotypic Complex Identify Specific Keratin Regions Involved in Intermediate Filament Assembly. Structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Hinbest AJ, Ho M, Eldirany SA, and Bunick CG (2020). Crystal structure of keratin 1/10(C401A) 2B heterodimer demonstrates a proclivity for the C-terminus of helix 2B to form higher order molecular contacts. Yale J Biol Med 93, In press. [PMC free article] [PubMed] [Google Scholar]
- Robert A, Hookway C, and Gelfand VI (2016). Intermediate filament dynamics: What we can see now and why it matters. Bioessays 38, 232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolova AV, Kreplak L, Wedig T, Mücke N, Svergun DI, Herrmann H, Aebi U, and Strelkov SV (2006). Monitoring intermediate filament assembly by small-angle x-ray scattering reveals the molecular architecture of assembly intermediates. Proc Natl Acad Sci U S A 103, 16206–16211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert PM, Marekov LN, Fraser RD, and Parry DA (1993). Keratin intermediate filament structure. Crosslinking studies yield quantitative information on molecular dimensions and mechanism of assembly. J Mol Biol 230, 436–452. [DOI] [PubMed] [Google Scholar]