Fig. 1. Histone mRNA metabolism.
A. Cell cycle regulation of histone mRNA. Histone mRNAs accumulate at high levels only in S-phase. When DNA synthesis is inhibited in S-phase, histone mRNAs are rapidly degraded. Histone mRNAs are also rapidly degraded at the end of S-phase, and histone mRNA biosynthesis is down-regulated.
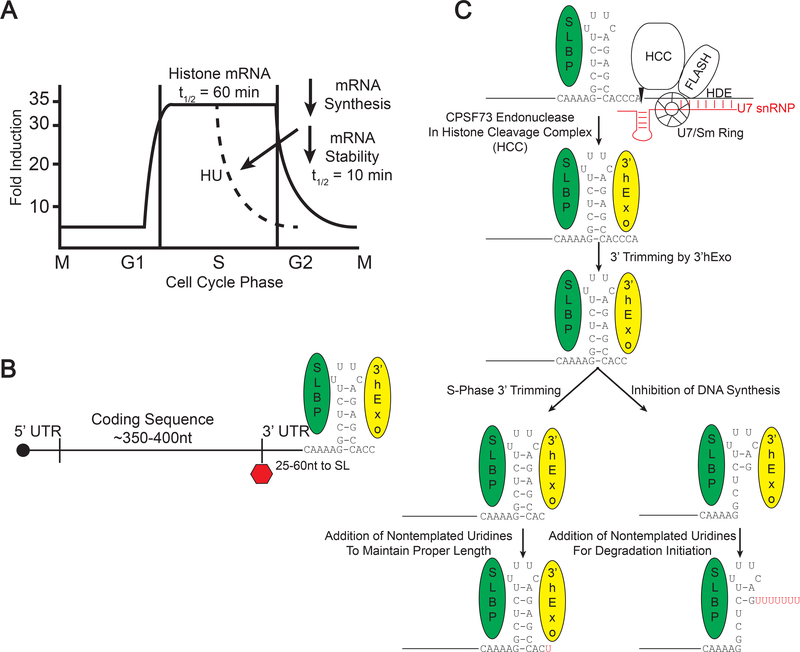
B. Structure of the histone mRNP. Histone mRNAs are capped and end in a conserved stemloop sequence at the 3’ end which binds to both SLBP and 3’hExo. C. Histone mRNA is formed by an endonucleolytic cleavage 5 nts after the stemloop, which requires SLBP, that binds the stemloop and U7 snRNP complexed with FLASH and the Histone Cleavage Complex (HCC) which contains the endonuclease CPSF73. After processing, 3’hExo binds and trims 2 nts from the 3’ end. In the cytoplasm during S-phase, there may be some trimming of the 3’ end, and the 3’ end is restored to 3 nts after the stemloop by uridylation. When DNA synthesis is inhibited, 3’hExo can degrade into the stemloop, where there are long U-tails added to the degradation intermediates.