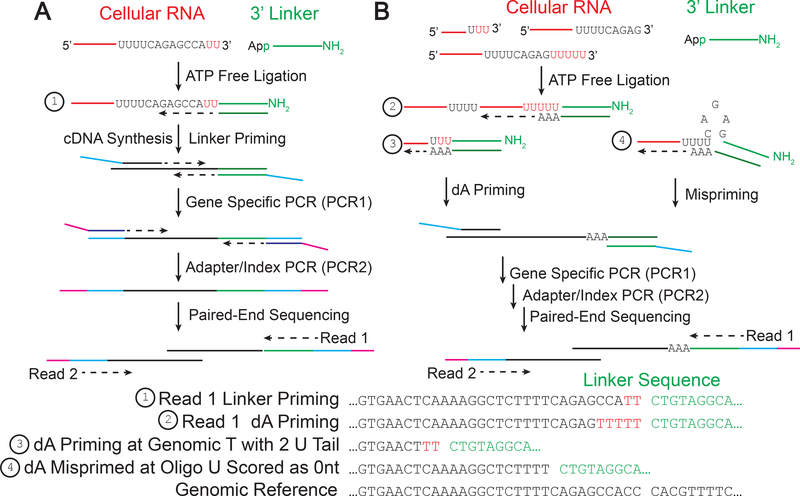
Fig. 2. Overview of the EnD-Seq/AppEnd workflow.
A. Total cellular RNA (or RNA from any subcellular fraction of interest) is ligated to a pre-activated synthetic linker sequence which is added to the 3’ OH on all RNAs. Note if the 3’ end is further modified (e.g. 2′Omethyl or phosphorylated) those ends will not be ligated. An oligonucleotide complementary to the linker is used to prime cDNA synthesis via reverse transcription. Specific sets of mRNAs can be amplified by PCR to specifically amplify the 3’ ends of the mRNAs of interest. The linker and gene specific primers are used to initially amplify the cDNA and a second round of PCR is used to add indices for each library and adapters for deep sequencing. Read 1 includes the linker sequence and 3’ terminus of the RNA and Read 2 includes the library indices and the gene sequence. The reads are aligned to a reference genome, the linker sequence is identified using the AppEnd algorithm [18]. Any nontemplated nucleotides on the 3’ termini (as small as one nt) will be identified as nts between the last template nucleotide and the start of the linker (example 1).
B. One can also select for specific non-templated nts (e.g. 3 uridines) at the 3’ end by adding 3 A’s to the 3’ end of the cDNA primer, followed by amplification of the cDNA for sequencing as in panel A. Note that this will allow one to determine the length of the oligo(U) tail (example 2). Note that if one primes at a template U or U’s, the AppEnd algorithm will identify only the nontemplated nts as part of the tail (example 3). We have observed mispriming at stretches of U’s, and these will be scored as no U tails (example 4). This method does an excellent job of identifying non-templated oligo(U) tails >2, and any shorter tails identified need to be interpreted cautiously.