Figure 4:
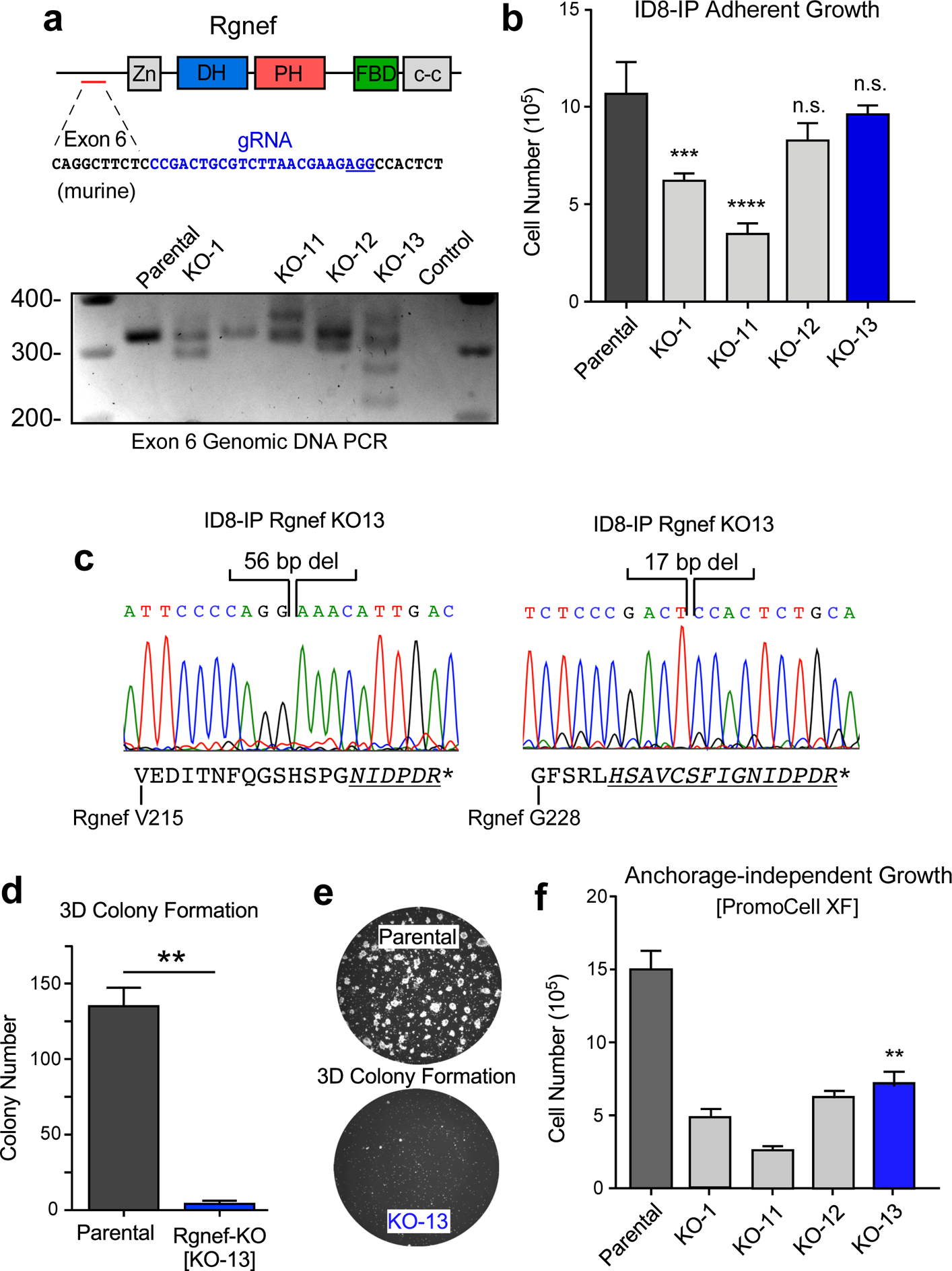
Rgnef loss specifically impairs 3D cell growth. (a) Schematic of CRISPR/Cas9 gRNA targeting of murine Rgnef exon 6 (blue text) within the N-terminal domain. Also shown are Rgnef protein domains: Zn - zinc finger, DH – Dbl homologous, PH - pleckstrin homology, FBD – FAK binding domain, c-c - coiled-coil region. Below: Exon 6 PCR was performed with genomic DNA to identify insertions or deletions in the indicated Rgnef KO clones. Parental ID8-IP cells yield a 393 bp band. (b) Adherent growth of ID8-IP and the indicated Rgnef-KO clones over 4 days (n.s.= no significance, *** P ≤0.001, **** P ≤ 0.0001, n=3 technical replicates, +/− SD). (c) Sanger sequencing of ID8-IP Rgnef KO-13 clone exon 6 identifying two independent deletions (56 and 17 bp) resulting in predicted translational termination. Alternative reading frame is underlined and italicized with position of stop codon (*) denoted (Uniprot protein G5E8P2). (d) ID8-IP and Rgnef KO-13 colony formation (** P ≤ 0.01, n=2 independent experiments, +/− SD). (e) Representative images of colonies in Matrigel. (f) Anchorage-independent growth of ID8-IP and the indicated Rgnef KO clones in serum-free conditions over 5 days (+/− SD, **P ≤ 0.01).
