Fig. 4.
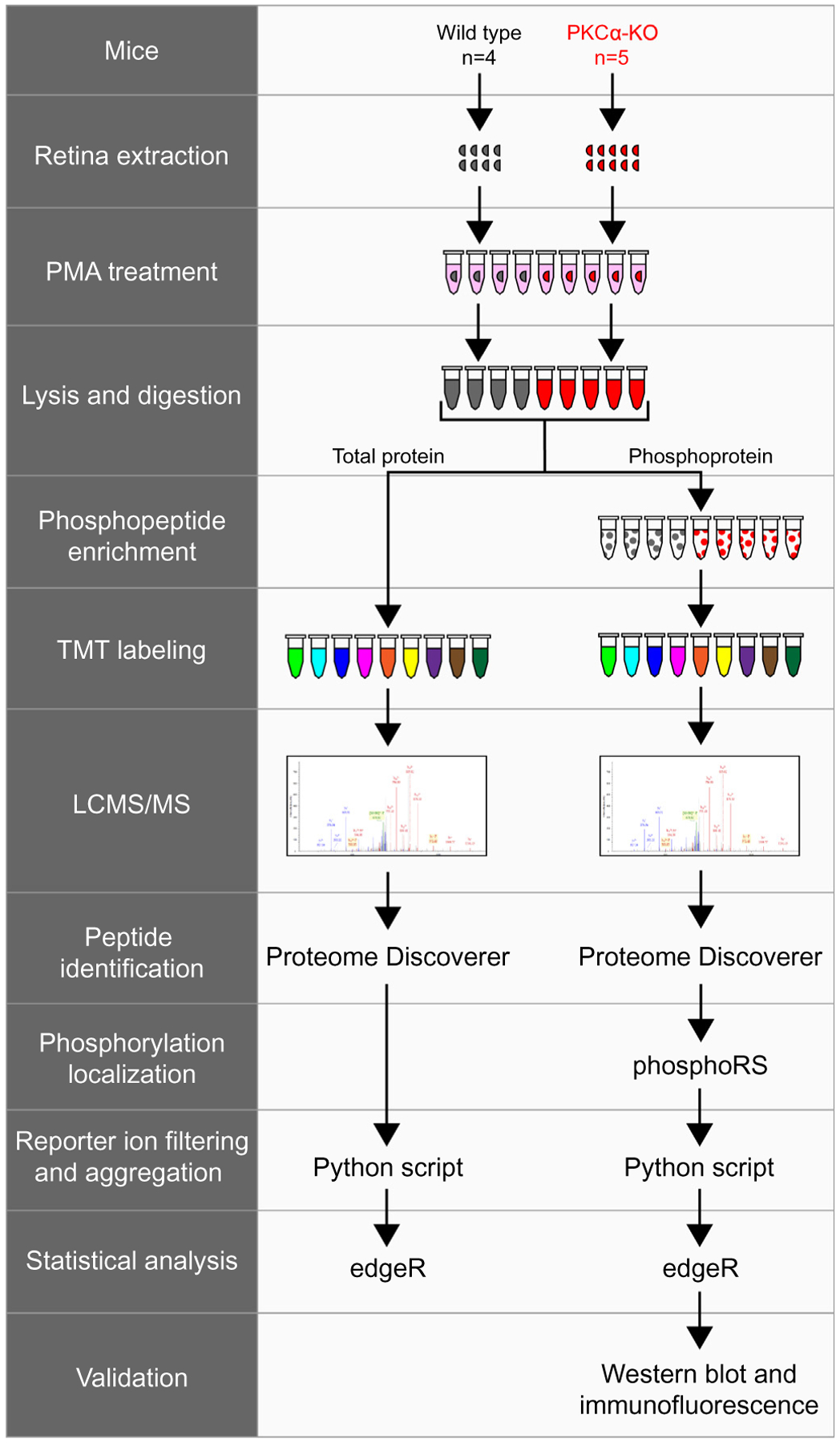
Experimental workflow of total protein and phosphopeptide identification. Wild type (n = 4) and PKCα knockout (KO) (n = 5) retinas were extracted and treated with PMA before lysis and trypsin digestion. A small fraction of each sample was removed for total protein analysis, while the rest of the samples underwent phosphopeptide enrichment. Following TMT labeling, samples were combined and analyzed by LCMS/MS. Tandem mass spectrometry data was collected on an Orbitrap Fusion and proteins were identified using Proteome Discoverer (SEQUEST and Percolator). Phosphorylation site localization was scored using phosphoRS, and reporter ion intensities were filtered and aggregated with an in-house Python script. TMT reporter ion intensities from total proteins or from phosphopeptides were tested for differential expression using the Bioconductor package edgeR. The presence of representative phosphoproteins was validated in the retina by western blot and confocal immunofluorescence microscopy. PMA: phorbol 12-myristate 13-acetate.
