Abstract
Background:
Improved animal models are needed to understand the genetic and environmental factors that contribute to food allergy.
Methods:
We assessed the development of allergic phenotypes in 16 Collaborative Cross (CC) mouse strains, as well as the classic inbred C57BL/6J, C3H/HeJ, and BALB/cJ strains. Female mice were sensitized to peanut intragastrically with or without Cholera toxin, then challenged with peanut by oral gavage or intraperitoneal injection, and assessed for anaphylaxis. Peanut-specific immunoglobulins and T cell cytokines were quantified and total numbers of Tregs, mast cells, and basophils were determined.
Results:
Eleven of the 16 Collaborative Cross mouse strains had allergic reactions to intraperitoneal peanut challenge, whereas only CC027/GeniUnc mice reproducibly experienced severe symptoms following oral food challenge (OFC). CC027/GeniUnc, C3H/HeJ, and C57BL/6J all mounted a Th2 response against peanut, leading to the production of IL-4 and IgE but only the CC027/GeniUnc mice reacted to OFC. Orally-induced anaphylaxis in CC027/GeniUnc mice was correlated with serum levels of Ara h 2 in circulation, but not correlated with allergen-specific IgE or MMCP-1 levels, indicating systemic allergen absorption is important for anaphylaxis through the gastrointestinal tract. Furthermore, CC027/GeniUnc mice, but not C3H/HeJ or BALB/cJ mice, can be sensitized in the absence of Cholera toxin and react upon OFC to peanut.
Conclusions:
We have identified and characterized CC027/GeniUnc mice as a strain that is genetically susceptible to peanut allergy and prone to severe reactions following OFC. More broadly, these findings demonstrate the untapped potential of the CC population in developing novel models for allergy research.
Keywords: Collaborative Cross, Food Allergy, Peanut Allergy, Anaphylaxis, Mouse Model, Ara h 2
Capsule Summary:
This study demonstrates the use of the genetically diverse Collaborative Cross which identified an improved, orally reactive mouse model of peanut allergy, to study the etiology of food allergy and the development of new therapies.
Introduction:
Food allergy is a potentially life-threatening disease characterized by IgE-mediated degranulation of mast cells and basophils upon allergen ingestion. Affecting 6% of children and 4% of the general population, food allergy is a growing public health concern, with peanut allergy present in at least 1% of the US population (1–3). Although many food allergies are outgrown before adulthood, peanut and tree nut allergies persist in roughly 80–90% of the affected population (4). Significant progress in food allergy research has occurred over the last 10 years, including the development of potential therapies (5–10), identification of improved diagnostic approaches (3), and discovery of underlying immunologic mechanisms driving food allergies (11, 12). However, critical knowledge gaps exist surrounding the etiology of peanut allergy, including genetic, microbial, and environmental influences.
The laboratory mouse has been the premier model organism for understanding complex human diseases, and developing therapies for a variety of diseases. Despite concerns about the translation of data from specific mouse strains to larger human health responses (13), there has been a growing appreciation for the role that genetic diversity between inbred mouse strains has in different outcomes within experimental models of human diseases (14, 15). In order to better leverage and identify the causal genetic variants driving such disease differences, a number of mouse genetic reference panels (GRPs) including the BxD panel (16), and the subsequently generated inbred Collaborative Cross (CC) (17), and outbred Diversity Outbred (DO) (18) have been developed. These resources, panels of diverse mice with well-characterized genetics have been used to (a) characterize the breadth of disease phenotypes that can be attributed to genetic variation; (b) define new models of disease phenotypes not found in the small pool of classic mouse strains used in standard studies; and (c) identify those polymorphic genes driving differential disease responses. Critically, such systems improve upon the utility and rigor of experimental models, ultimately making them more relevant for modeling diverse human disease responses.
Since peanut allergy within the human population is a heritable (i.e. genetically influenced) trait, (19, 20) we sought to utilize the high levels of genome-wide genetic diversity present in the CC mice to improve our understanding of peanut allergy and its contributing factors. Numerous murine models are currently in use by our group and many others to study mechanisms and treatments of peanut allergy (21–23). However, these models often require powerful Th2-skewing adjuvants (e.g. Cholera toxin (24), Staphylococcal Enterotoxin B (25), or Aluminum hydroxide (26)) to sensitize animals, intraperitoneal (IP) challenge to elicit a reaction (24, 27), or complex modifications such as humanization (28–30). In a model commonly used by our group and others, C3H/HeJ mice are sensitized by weekly oral gavage of peanut extract and Cholera toxin and challenged by intraperitoneal (IP) injection with peanut extract (24). Importantly, while some reports demonstrate reactions upon oral challenge in the C3H/HeJ model described above (23), other groups, including our own, have not been able to successfully reproduce these findings (29, 31). A model that can both be sensitized and reproducibly react orally would allow for the study of therapies that alter the immune system in the gastrointestinal tract such as oral immunotherapy (OIT), as well as genetic and environmental factors driving these severe allergic reactions in a more physiologically relevant model of human disease. Here we report our screen of CC strains to identify orally-induced peanut anaphylaxis, the characterization of the peanut-specific immunologic responses, and novel insights into the anaphylactic reactions in mice through the GI tract. In concordance with prior assessment in the human population, we identified strong genetic control of the propensity to experience anaphylaxis after sensitization. We also identified the CC strain CC027/GeniUnc as a mouse strain that develops anaphylaxis following oral sensitization and challenge, even in the absence of the Th2-skewing Cholera toxin.
Methods:
Mice
CC mice were purchased from the UNC Systems Genetics Core (32). C57BL/6J, C3H/HeJ, and BALB/cJ mice were obtained from colonies maintained for less than five generations by the Pardo-Manuel de Villena lab from mice purchased from The Jackson Laboratory. All mice were bred at UNC, raised on standard mouse chow free of any peanut-containing ingredients, kept on a 12:12 light:dark cycle and transferred for sensitization at 4–6 weeks of age. Female mice were weaned into cages at a common cage density (between 3–5 mice/cage depending on the experiment), but with a diverse set of strains within each cage. In this way, effects of cage density and other cage-specific effects were removed from these studies. Throughout these studies, where possible, experimenters were blinded to the mouse strains being studied. All mouse work was conducted in compliance with UNC IACUC protocol 16–045 and 17–286.
Reagents
Peanut extract was created by mixing peanut flour (12% fat light roast, 50% protein; Golden Peanut Co.) in a 1:5 (wt:vol) ratio of phosphate buffered saline (PBS) with 1 mol/L NaCl and the soluble fraction was filter-sterilized as described previously (27). Protein concentration was determined by bicinchoninic acid assay (Pierce, Rockford, IL). Peanut extract was run on a NuPage gel to identify and compare relative quantities of peanut allergens before using.
Sensitization with Peanut plus Cholera Toxin and Challenge
4–6 week old female mice underwent weekly sensitization with 2 mg peanut extract and 10 μg Cholera toxin (List Biological laboratories, Campbell, CA) in 200 μL volume for three weeks by oral gavage followed by 1 week of 5 mg peanut extract and 10 μg Cholera toxin by oral gavage. One week after this final sensitization, mice were bled by submandibular bleed to collect serum for immunoglobulin quantification. The following day, mice undergoing an oral challenge were gavaged with 9 mg peanut extract while mice undergoing IP challenge received 200 μg peanut extract. Core body temperatures were monitored every fifteen minutes using a rectal thermometer (Physitemp, Clifton, NJ). For serum MMCP-1, histamine, and Ara h 1, 2, and 3 measurements, blood was collected 60 min after oral challenge. Serum levels of MMCP-1 (eBioscience, San Diego, CA), histamine (Beckman Coulter, Brea, CA) and Ara h 1, 2, and 3 (Indoor Biotechnologies, Charlottesville, VA) were measured by ELISA according to manufacturer’s instructions
Sensitization with Peanut in the Absence of Cholera Toxin and Challenge
4–6 week old female mice underwent sensitization for three weeks with either PBS once per week, 2 mg peanut extract once per week, 2 mg peanut extract three times per week, or 2 mg peanut extract plus 10 μg Cholera toxin once per week. All peanut groups received one final week of 5 mg peanut extract with or without Cholera toxin, at the frequency described. Bleeding and oral challenges were consistent with the procedure described above.
Immunoglobulin Quantification
For peanut-specific IgE (PNsIgE), PNsIgG1, and PNsIgG2a/c quantification, plates were coated with 20 μg/mL peanut extract diluted in carbonate-bicarbonate buffer (Sigma Aldrich, St Louis, MO). Samples were assayed on plates at 1:100, 1:20,000, and 1:1250, respectively. Ara h 1-specific IgE (sIgE), Ara h 2-specific IgE (Ah2sIgE), and Ara h 3-specific IgE (Ah3sIgE) plates were coated with 5 μg/mL of the appropriate purified peanut component diluted in carbonate-bicarbonate buffer. Samples were plated at a 1:20 dilution. IgE plates were all detected using the following antibodies in sequence: sheep anti-mouse IgE (0.5 μg/mL; Binding Site, Birmingham, UK), biotinylated donkey anti-sheep IgG (0.5 μg/mL; Accurate Chemical, Westbury, NY), neutravidin-horseradish peroxidase (HRP; 0.2 μg/mL; Pierce). IgG1 and IgG2a/2c ELISAs were detected with HRP-conjugated goat anti-mouse IgG1 (Southern Biotech, Birmingham, AL) or HRP-conjugated goat anti-mouse IgG2a (Southern Biotech, Birmingham, AL) and HRP-conjugated goat anti-mouse IgG2c (Southern Biotech, Birmingham, AL) respectively. Sure Blue TMB Microwell Peroxidase Substrate and Stop Solution (KPL, Gaithersburg, MD) were applied to all plates. Plates were read on an Epoch Microplate Spectrophotometer (BioTek Instruments, Winooski, VT). Total IgE was analyzed by ELISA, (Affymetrix, Santa Clara, CA) and run according to manufacturer’s instructions. All ELISA data was analyzed using Gen5 software.
mRNA and Cytokine Protein Quantification
Spleens were collected from both naïve as well as peanut-sensitized mice 1 week after oral challenge. mRNA abundance levels were quantified using real-time PCR and Sybr green methodology, as previously described (33). Briefly, total RNA was extracted using RNA kits (Qiagen, Germantown, MD). Reverse transcription was performed using random decamers as primers and Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). The abundance of resultant mRNA-derived cDNA was determined by qRT-PCR analysis, using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, Hercules, CA), primers specific for genes of interest (See Table S1 for sequences), and a StepOne Plus cycler (Applied Biosystem, Foster City, CA). Each set of primers spans at least two introns so that contaminating genomic DNA either cannot be amplified due to large product size or can be easily identified based on its size on agarose gel. Primers for 18S rRNA were obtained from Ambion (Austin, TX). The specificity of each real-time PCR target was confirmed by melt temperature analysis and agarose gel fragmentation of amplicons. To quantify mRNA abundance, a standard curve for each target mRNA, as well as for 18S rRNA, was generated from serial dilutions of cDNAs derived from a pooled intestinal cDNA library. The relative abundance of mRNA of interest in each sample was determined based on its corresponding standard curve, and normalized against the abundance of 18S rRNA. For protein analysis, splenocytes were isolated and cultured for 96 hours in the presence of 200 μg/mL peanut extract. Supernatants were collected and run on Meso Scale Discovery plates to determine levels of IL-4, IL-5, IL-13, TNF-α, IFN-γ, IL-12p40, IL-10 according to manufacturer’s instructions (MSD, Rockville, MD).
Flow Cytometry
For analysis of Regulatory T (Treg) cells, splenocytes were collected 1 week after oral challenge. Splenocytes were stimulated with 200 μg/mL peanut extract for 7 days. Tregs were then labeled using FITC rat anti-mouse CD4 Clone RM4–5 (BD Bioscience, San Diego, CA), PE anti-mouse/rat/human FoxP3 Clone 150D (Biolegend, San Diego, CA), and APC rat anti-mouse CD25 Clone PC61 (BD Biosciences, San Diego, CA). For determination of basophil levels, whole blood was collected by submandibular bleed 1 week after sensitization. Cells were stained with anti- mouse IgE FITC Clone 23G3 (eBioscience, San Diego, CA), PerCP/Cy5.5 anti-mouse CD49b Clone DX5 (Biolegend, San Diego, CA), PE anti-mouse CD200R Clone OX-110 (Biolegend, San Diego, CA). Flow Cytometry was performed on a Beckman Coulter CyAn ADP and analyzed using FlowJo (v10). Basophils were gated as IgE+CD49b+ and expressed as a percentage of lymphocytes. Tregs were gated as CD4+CD25+FoxP3+ and expressed as a percentage of CD4+ lymphocytes.
Histology
Proximal jejunum was harvested from peanut-sensitized mice 1 week after challenge, and fixed with cold 4% paraformaldehyde in phosphate-buffered saline overnight and paraffin-embedded. Cross-cut sections (at a thickness of 7 μm) were subjected to immunostaining with an antibody specific for mast cell tryptase (1:180, Abcam, ab151757) or for CD117 (c-kit, 1:150, ThermoFisher, PA5–16770). Antibody-antigen complexes were detected using an ABC kit (Vector Laboratories, Burlinggame, CA) and visualized by incubation with DAB (Vector Laboratories or Sigma-Aldrich, St Louis, MO). For mast cell tryptase-positive cell quantification, immunostained sections were then subjected to counterstaining for cell nuclei with 0.1% methylene blue in acetic acid. To estimate the number of mast cell tryptase-positive cells in mucosa, 2 – 4 villi and the crypts under the villi were randomly selected, mast cell tryptase-positive cells with clear nuclei within delineated villi and crypts were counted. Total number of cells was determined by counting methylene blue-stained cell nuclei, and the percentage of mast cell tryptase-positive cells were calculated. For each mouse, 850–1,640 cells were counted.
Statistical analysis
GraphPad/Prism version 7.02 was used to analyze all data. Mann-Whitney U, Spearman Correlation, and Unpaired-t tests were performed and a p-value <0.05 was considered significant. For cytokine protein level, values at or below the lower limit of detection were assigned half of the value of the lower limit of detection for that particular MSD assay.
Results:
CC027/GeniUnc female mice react severely to both oral and IP challenge with peanut extract
To assess the role that genetic variation plays in controlling anaphylaxis following sensitization with peanut allergen, female mice from 16 CC strains were screened using an established sensitization model (27) (Figure 1). These CC strains were chosen based on the fact that their well-characterized genetic makeup is representative of the larger CC population (34), as well as prior reports (14, 35) of aberrant disease present in specific strains. All mice underwent the same four-week sensitization regimen followed by half of the mice in each strain receiving a 200 μg peanut extract challenge via IP injection and the other half receiving a 9 mg peanut extract challenge via oral gavage. Following either oral food challenge (OFC) or IP challenge, CC strains were grouped into three types of reactors: Strains that do not react regardless of challenge route (Figure 2A: OFC, Figure 2D: IP); strains that reacted mildly (mean body temperature decreases between 1.5–3°C; Figure 2B: OFC, Figure 2E: IP), and strains that reacted severely (mean body temperature decreases > 3°C; Figure 2C: OFC, Figure 2F: IP). As expected, the screen identified many more mild and severe reactors following IP challenge than following oral challenge, however responses were highly concordant across routes of challenge (i.e. if a strain was a non-reactor in the IP cohort, it was also a non-reactor in the OFC cohort). Two strains, CC027/GeniUnc (referred to as CC027 in figures) and CC012/GeniUnc, were classified as strong reactors following OFC, and were also classified as strong reactors to IP challenge with peanut extract, suggesting that they could be potential models for severe anaphylaxis following peanut sensitization.
Figure 1.
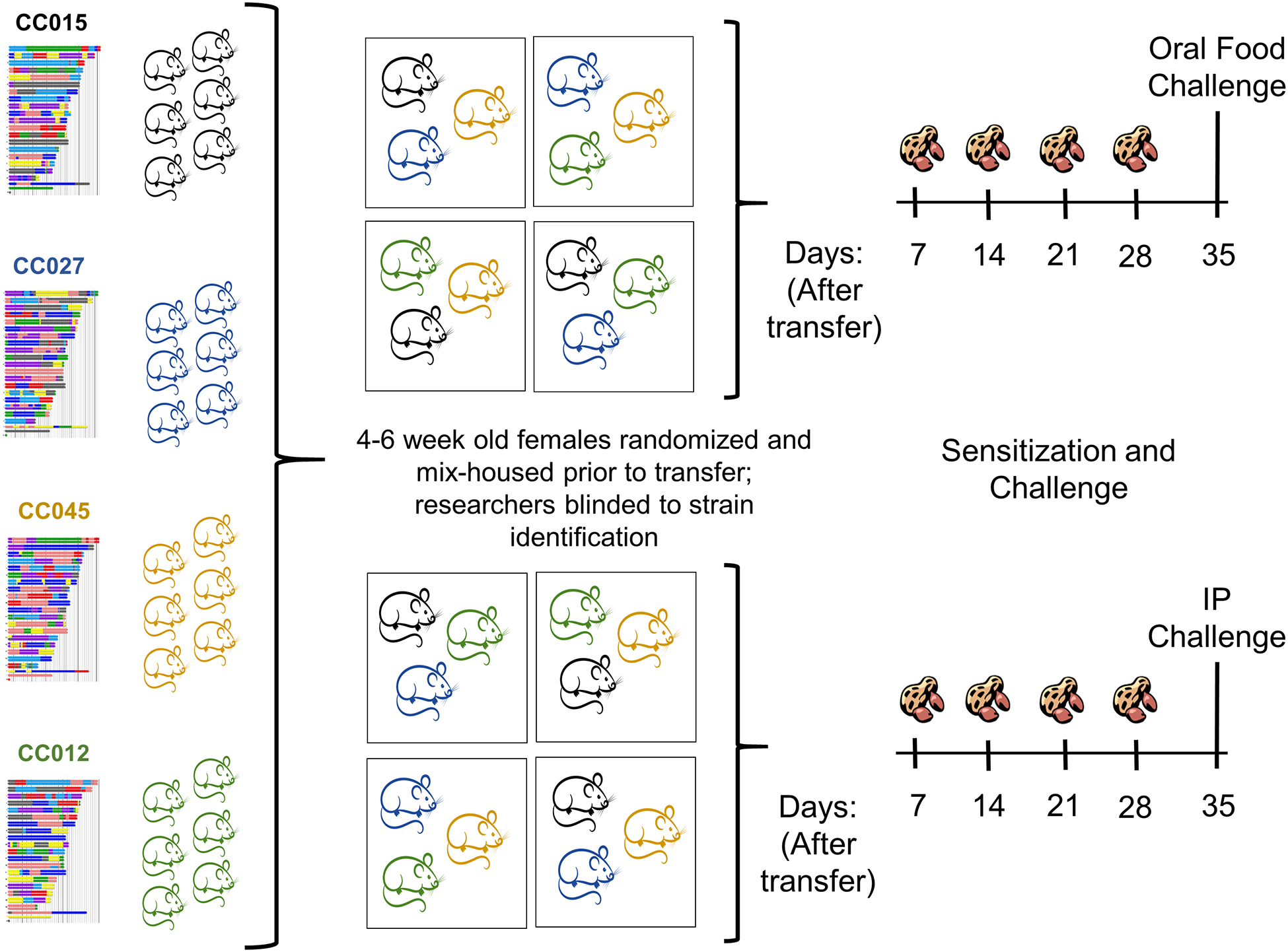
Collaborative Cross screening approach. Schematic shows 4 representative strains of the 16 strains screened. Six female mice between the ages of 4 and 6 weeks from each strain were mixed so that each cage contained 3–5 mice from different strains. Mice were then transferred from the UNC Systems Genetics Core to the UNC Food Allergy Initiative where researchers were blinded to the identification of each strain. Mice were sensitized intragastrically with peanut extract and Cholera toxin for 4 weeks before undergoing either an OFC (n=3/strain) or IP challenge (n=3/strain) with peanut extract.
Figure 2.
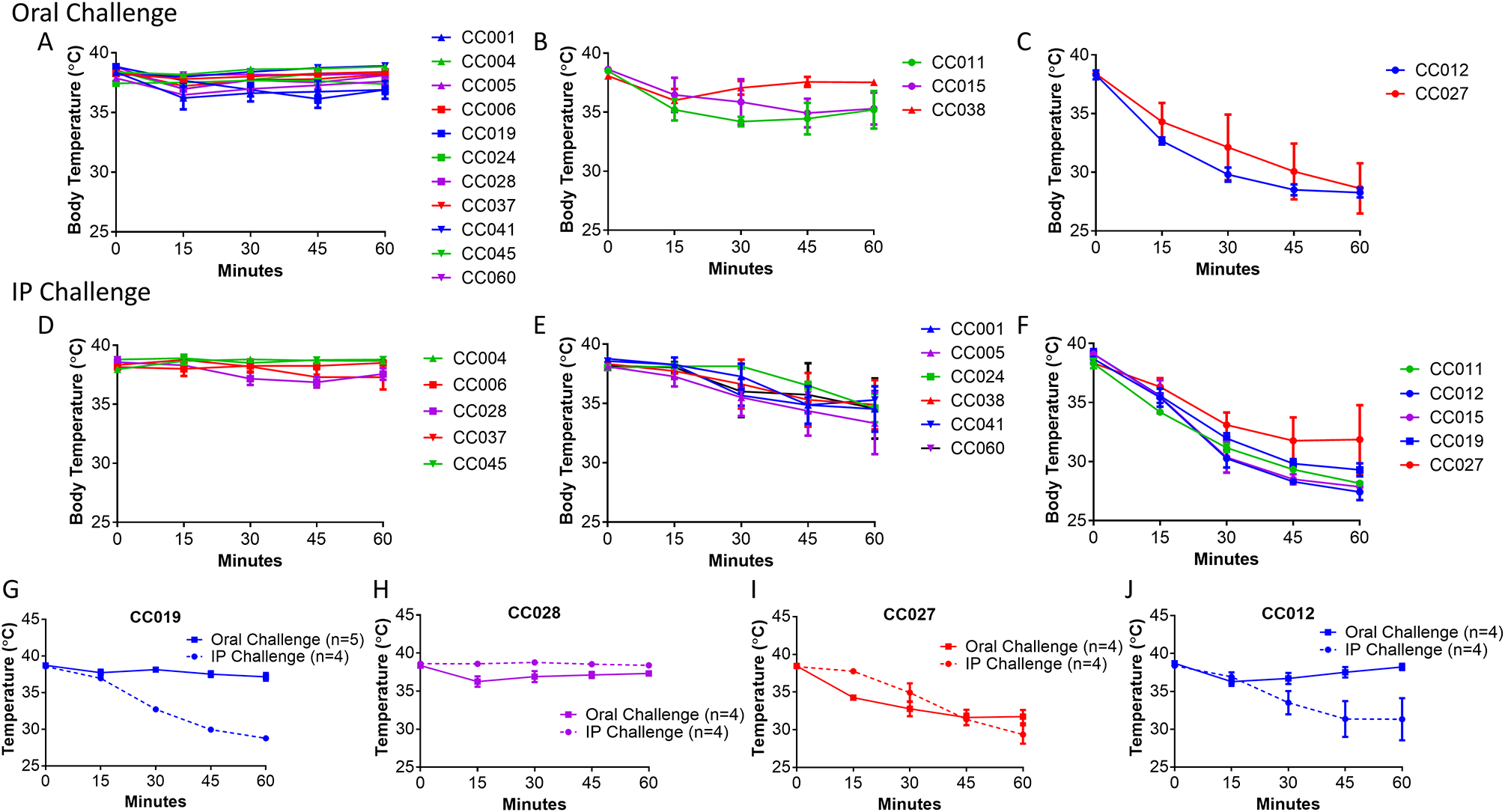
Anaphylaxis in peanut-sensitized Collaborative Cross strains following peanut challenge. Oral and IP challenges revealed Collaborative Cross strains that are non-reactors (A, D), mild reactors (B, E) and severe reactors (C, F) as measured by decreased body temperature. Challenges were repeated with an IP reactor control (G), a non-reactor control (H), and oral reactors (I, J).
In order to validate the findings of this initial screen, we conducted a second experiment with the OFC severe responders CC027/GeniUnc, CC012/GeniUnc, as well as the non-responder CC028/GeniUnc and IP-only responder CC011/Unc. While the results of this experiment were largely concordant with the initial screen (Fig 2G–J), only CC027/GeniUnc exhibited a severe reaction following OFC. We therefore concluded that CC027/GeniUnc represents a robust OFC-reaction model derived from our screen of the CC.
CC027/GeniUnc, but not C3H/HeJ, C57BL/6J, or BALB/cJ, react on oral challenge despite all making IgE to peanut allergens
Immune responses of sensitized CC027/GeniUnc females were compared to those of female mice from the classical inbred C3H/HeJ, C57BL/6J, and BALB/cJ strains. All four strains were sensitized using the previously described 4 week sensitization schedule and then underwent an OFC with 9 mg peanut extract. Consistent with our previous experiments, CC027/GeniUnc mice experienced severe systemic reactions with body temperatures decreasing over the course of 60 min following OFC (Figure 3A), whereas C3H/HeJ, C57BL/6J, and BALB/cJ mice had essentially no change in body temperature following OFC. These results confirm the utility of CC027/GeniUnc as an orally reacting allergy model. Despite only CC027/GeniUnc mice reacting upon OFC, C3H/HeJ and C57BL/6J make peanut-specific IgE (PNsIgE), peanut-specific IgG1 (PNsIgG1), and peanut-specific IgG2a/c (PNsIgG2a/2c) as well as IgE to the major peanut components, Ara h 1, Ara h 2, and Ara h 3 (Figure 3B–G). After sensitization, CC027/GeniUnc make significantly more PNsIgE than C3H/HeJ (p<0.05) but not C57BL/6J (Figure 3B) or BALB/cJ (Figure E1). PNsIgG1 levels are not different between CC027/GeniUnc, C3H/HeJ and C57BL/6J (Figure 3C) while PNsIgG2a/c levels are higher in CC027/GeniUnc than C57BL/6J but not different from C3H/HeJ (Figure 2D; p<0.01). CC027/GeniUnc mice also had significantly more total IgE than C3H/HeJ (p<0.01) but not significantly different total IgE levels from C57BL/6J after sensitization (Figure 3H).
Figure 3.
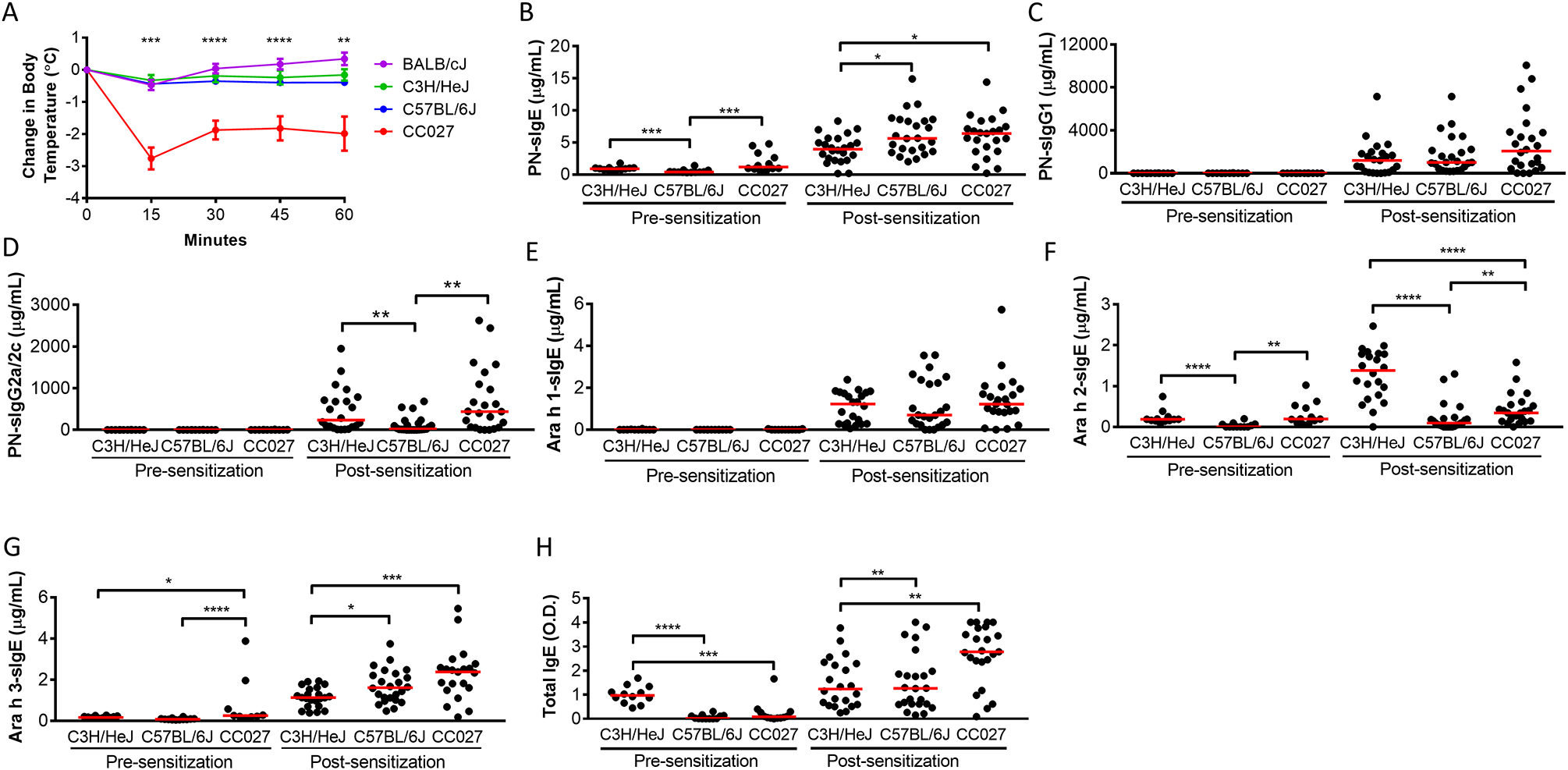
Immune response of CC027/GeniUnc to peanut extract relative to that of C3H/HeJ, C57BL/6J, and BALB/cJ mice. CC027/GeniUnc is represented as CC027 in figures. Body temperatures following oral challenge with peanut extract (n≥12/strain; representative of 3 independent experiments) (A). Serum levels of immunoglobulins following 4 weeks of sensitization (B-H). Mann-Whitney U Test *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Statistical significance represents comparisons of C3H/HeJ, C57BL/6J, and BALB/cJ relative to CC027/GeniUnc (A).
Given that PNsIgE and PNsIgG1 levels were different between CC027/GeniUnc and at least one of the two classical inbred strains, we assessed whether PNsIgE or PNsIgG1 levels within CC027/GeniUnc mice were correlated with anaphylaxis reaction severity. We found that PNsIgE and PNsIgG1 levels did not correlate with reaction severity in CC027/GeniUnc mice and thus do not explain the increased reactivity of these mice (Figure E2A–B). Together, these data show that CC027/GeniUnc make immunoglobulins to peanut and peanut components, but that the strain-specific production of immunoglobulins alone does not distinguish CC027/GeniUnc from C3H/HeJ or C57BL/6J.
CC027/GeniUnc mounts a Th2 cellular response with little Th1 or regulatory cytokine response to peanut
Secreted cytokines from peanut-stimulated splenocytes were quantified for CC027/GeniUnc, C3H/HeJ, and C57BL/6J mouse strains to determine T cell phenotypes. All three strains produce IL-4 and levels are not significantly different across the strains (Figure 4A). IL-12-p40 was significantly elevated in C3H/HeJ relative to both C57BL/6J and CC027/GeniUnc (Figure 4F). For the remaining five cytokines (IL-5, IL-13, TNF-α, IFN-γ, IL-10), we found that CC027/GeniUnc had significantly lower levels than either C57BL/6J or C3H/HeJ (Figure 4B–E, H). The ratio of IL-4 to IFN-ɣ, which is indicative of a Th2-skewed response [23–25] was higher in CC027/GeniUnc compared to C3H/HeJ or C57BL/6J (Figure 4G). These secreted cytokine data demonstrate that CC027/GeniUnc mice mount Th2 responses to peanut antigen with limited production of IFN-ɣ, IL-12p40, TNF-α, and IL-10.
Figure 4.
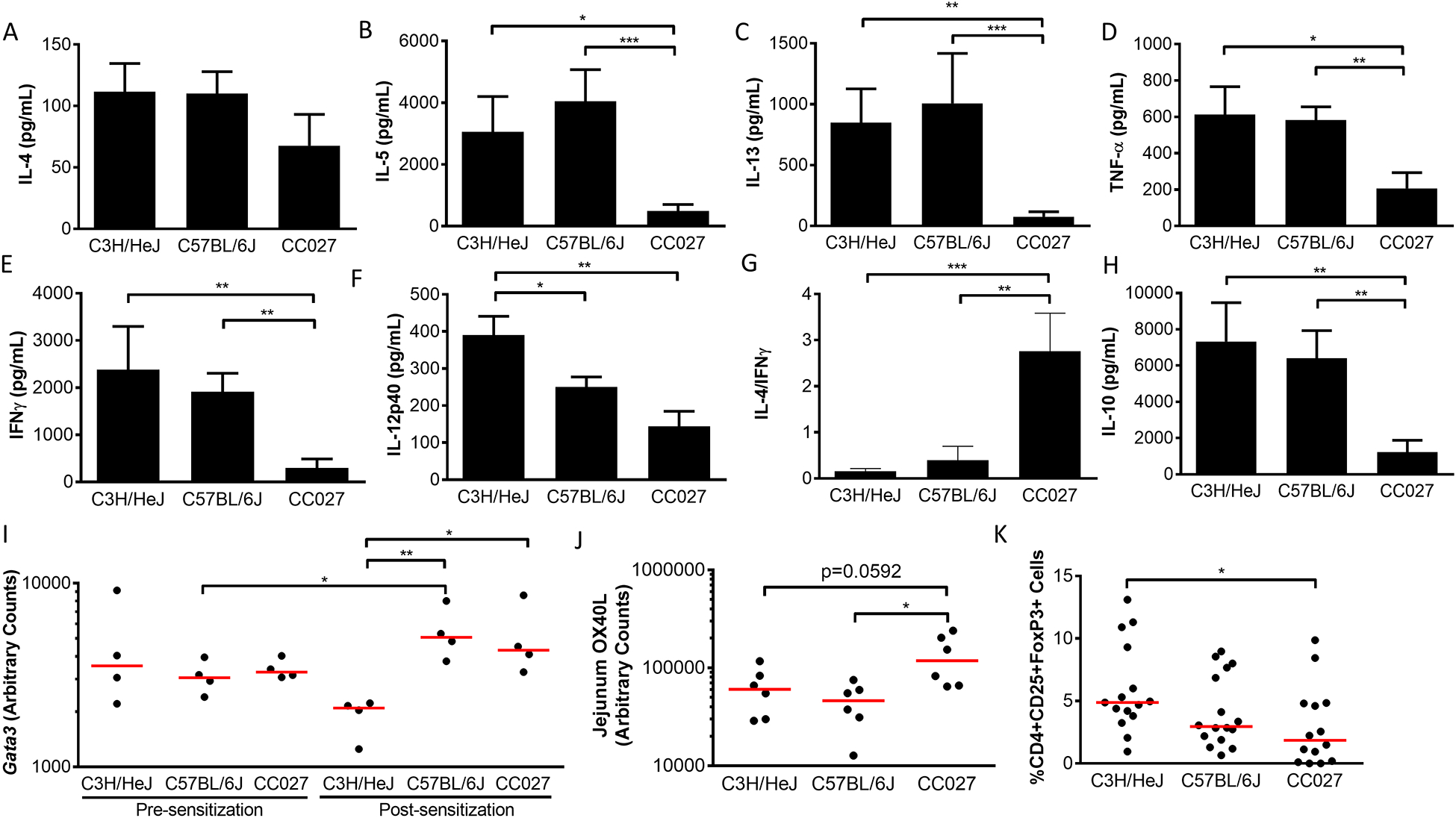
Cellular responses in CC027/GeniUnc, C3H/HeJ, and C57BL/6J. Splenic cytokines 96 hours following peanut-stimulation (n=10/strain) (A-H), mRNA expression (I-J), and CD4+CD25+FoxP3+ regulatory T cells (K) 1 week following oral challenge. Mann-Whitney U Test *p<0.05, **p<0.01, ***p<0.001 (A-H; K); unpaired t-test *p<0.05, **p<0.01 (I-J).
Concurrent with the finding that CC027/GeniUnc mice show a Th2 response to peanut, we found that CC027/GeniUnc have higher levels of Gata3 mRNA than C3H/HeJ (p<0.05), albeit with similar levels to C57BL/6J (Figure 4I). CC027/GeniUnc also had a lower T-cell regulatory response as indicated by reduced CD4+CD25+Foxp3+ Treg levels (Figure 4K) and decreased IL-10 protein production than the classical inbred strains (p<0.01; Figure 4H).
Sensitization-induced changes in mRNA expression levels were also analyzed for a few selected genes. Interestingly, CC027/GeniUnc have lower expression levels of Il10 and Il12 after sensitization than they do at baseline (p<0.05; Figure E3A–B), suggesting that sensitizing these animals results in decreased production of regulatory and Th1-type cytokines. However, sensitization did not change Il10 or Il12 mRNA levels in either C3H/HeJ or C57BL/6J (Figure E3A–B). Other reports have shown that Ox40L expression increases in dendritic cells following sensitization (36). We found Ox40L mRNA levels in the small intestine to be increased in CC027/GeniUnc compared to C3H/HeJ (p=0.0592) and C57BL/6J (p<0.05) showing an additional effect of sensitization in these mice (Figure 4J). Overall, T cell responses appear to favor pro-allergic responses to peanut with the presence of Th2 cytokines and limited Th1 cytokine production, lower numbers of Tregs, and less regulatory cytokine IL-10.
Effector cells are more prevalent in CC027/GeniUnc mice than classic inbred strains
Basophils and mast cells are the two main effector cells implicated in allergic reactions to foods (37). We quantified basophil frequency in blood after sensitization using flow cytometry. CC027/GeniUnc had an increased percentage of IgE+CD49b+ basophils circulating after sensitization compared to C3H/HeJ (p<0.05) and C57BL/6J (p<0.01; Figure 5A). Furthermore, the CC027/GeniUnc basophils also had less of the inhibitory receptor, CD200R1 than the other two strains (p<0.001; Figure 5B) (38). Tissue samples of the small intestine were stained for tryptase+ mast cells. CC027/GeniUnc had an increased percentage of tryptase+ cells, suggesting increased mast cell presence in the tissue (Figure 5C–D). Similar findings were noted with toluidine blue staining of mast cells in tissue (Figure E4). Taken together, CC027/GeniUnc have an increased number of basophils in circulation that may lack negative feedback mechanisms driven by CD200R, and also an excess of mast cells in the GI tract.
Figure 5.
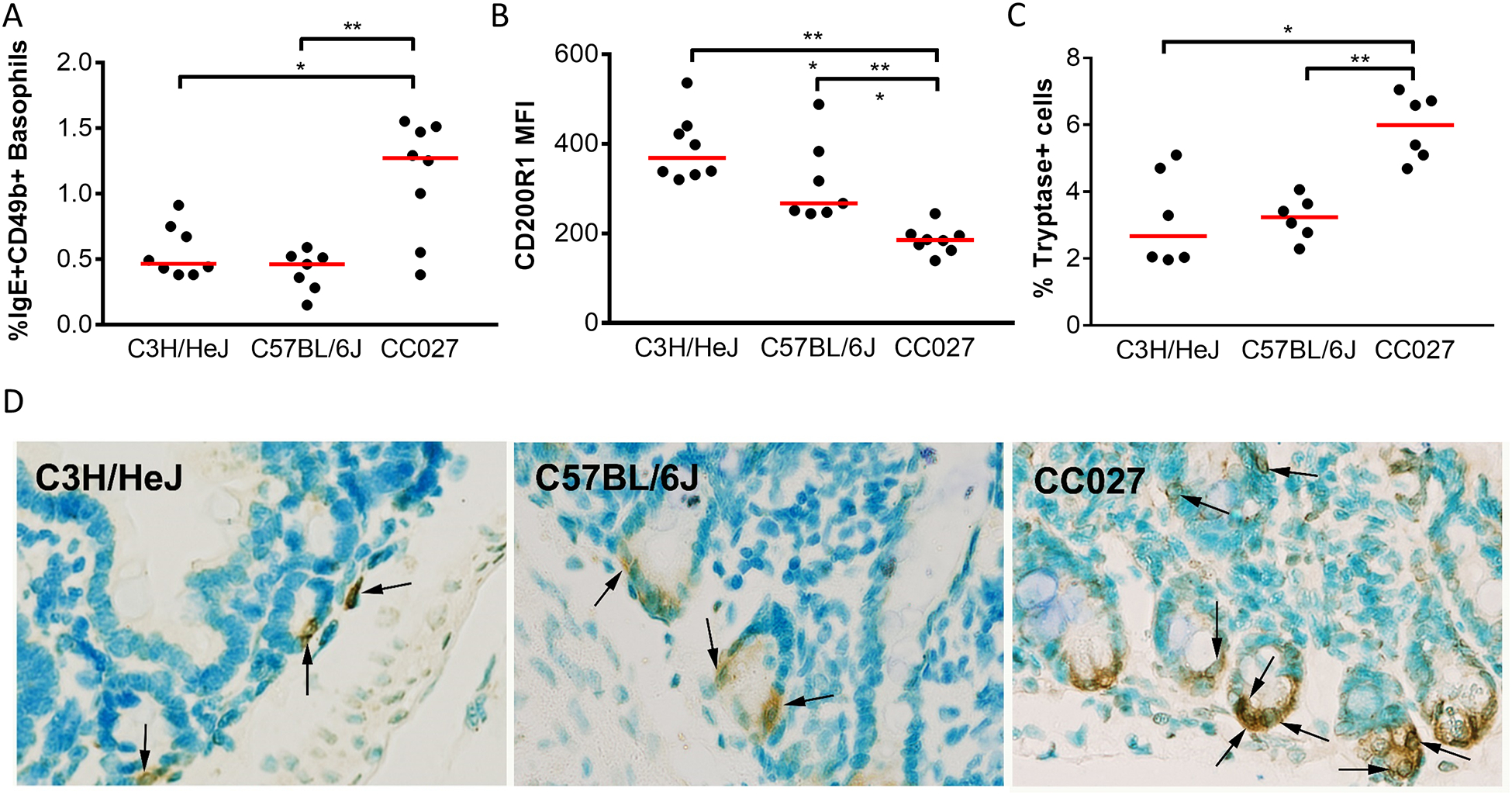
Enumeration of effector cells in CC027/GeniUnc, C3H/HeJ, and C57BL/6J. Percent IgE+CD49b+ basophils (A) and basophil inhibitory receptor, CD200R1 in whole blood (B). Jejunal tryptase+ mast cells quantified 1–3 weeks following challenge (C) and representative staining images shown with arrows indicating tryptase+ cells (D). Mann-Whitney U Test *p<0.05, **p<0.01, ***p<0.001.
Reaction severity in CC027/GeniUnc correlates with serum levels of Ara h 2 but not MMCP-1 during oral challenge
To further characterize the severe reaction observed in CC027/GeniUnc mice, blood was collected from the mice 60 min following OFC. Serum levels of mucosal mast cell protease-1 (MMCP-1), a mediator released by degranulated mast cells in the gastrointestinal tract was measured to verify that mast cell degranulation could be detected in the reacting animals. Serum MMCP-1 was detectable in both C3H/HeJ and CC027/GeniUnc, but not C57BL/6J (Figure 6A), though only CC027/GeniUnc showed signs of a systemic reaction. Histamine levels were also quantified following OFC and were not different between strains (Figure E5A). Within CC027/GeniUnc mice, serum levels of MMCP-1 were not correlated with reaction severity (Figure 6B; Spearman r=0.2196, p=0.4109); histamine levels were also not correlated with reaction severity (Figure E5B). Histamine levels in serum 5 minutes post-OFC were also not different between strains and did not correlate with reaction severity (data not shown). Concurrently, serum levels of the major peanut allergen, Ara h 2, were measured 60 min post-challenge by ELISA to determine the amount of allergen being absorbed into the blood stream. CC027/GeniUnc had significantly higher levels of Ara h 2 in serum, compared to C57BL/6J (p<0.05) and C3H/HeJ (p<0.0001; Figure 6C). Interestingly, Ara h 2 quantity positively correlates with reaction severity in CC027/GeniUnc mice (Figure 6D; Spearman r=0.69, p=0.0028). Two other major peanut allergens, Ara h 1 and Ara h 3, were not detectable in serum post-challenge.
Figure 6.
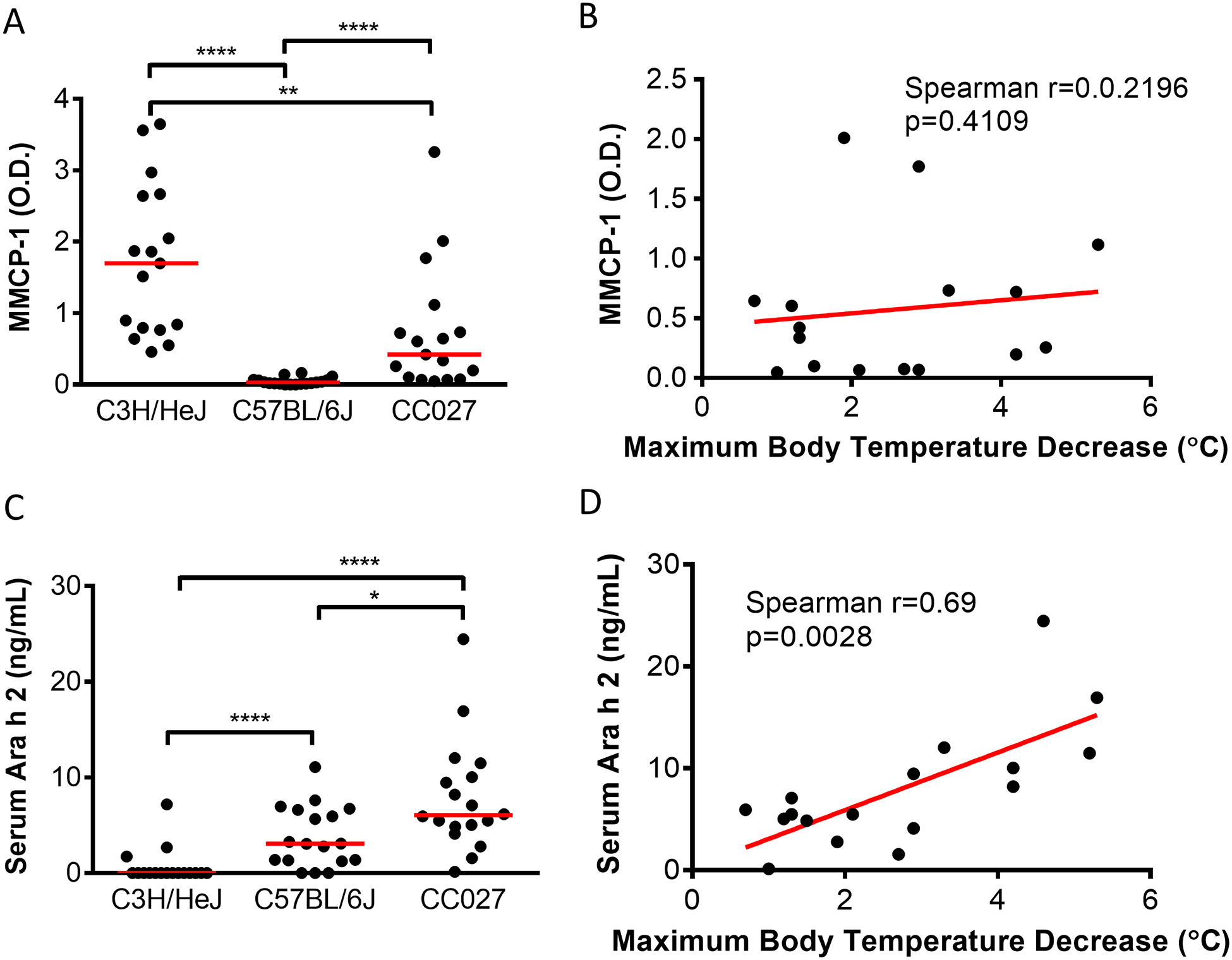
Post-OFC serum levels of mast cell degranulation marker and the major peanut allergen Ara h 2. Serum levels of MMCP-1 (A) and Ara h 2 (C) 60 min after oral challenge; Mann-Whitney U Test *p<0.05, **p<0.01, ****p<0.0001. Correlations between MMCP-1 (B) or Ara h 2 (D) and maximum body temperature decrease following oral challenge in CC027/GeniUnc.
CC027/GeniUnc mice can be sensitized in the absence of Cholera toxin and react upon oral challenge with peanut extract
Given the oral reactivity of the CC027/GeniUnc female mice, we then tested whether they could be sensitized with peanut extract alone (i.e. without Cholera toxin or any additional adjuvant) in comparison with C3H/HeJ and BALB/cJ mice. Following oral challenge with peanut, all groups of C3H/HeJ mice and BALB/cJ did not experience decreased body temperatures (Figure 7A and B). However, CC027/GeniUnc mice sensitized with peanut alone, once or three times per week, experienced on average a greater than 3 and 4°C drop, respectively, comparable to mice sensitized with peanut plus Cholera toxin (Figure 7C). All strains had increased PNsIgE after sensitization with peanut plus Cholera toxin. CC027/GeniUnc mice sensitized with peanut three times per week had higher levels of PNsIgE, PNsIgG1 and PNsIgG2a post-sensitization compared to C3H/HeJ (p<0.05), but not BALB/cJ (Figure 7D, S6A and B). CC027/GeniUnc mice sensitized three times a week with peanut had increased PNsIgE compared to the group sensitized with PBS (p<0.01), while mice sensitized with peanut plus Cholera toxin had higher PNsIgE (p<0.0001). There was no difference in PNsIgE between CC027/GeniUnc mice sensitized with peanut alone three times or once per week. Overall, these data demonstrate that CC027/GeniUnc mice can be sensitized with peanut in the absence of Cholera toxin, as evidenced by their decreased body temperature after OFC and increased levels of PNsIgE.
Figure 7:
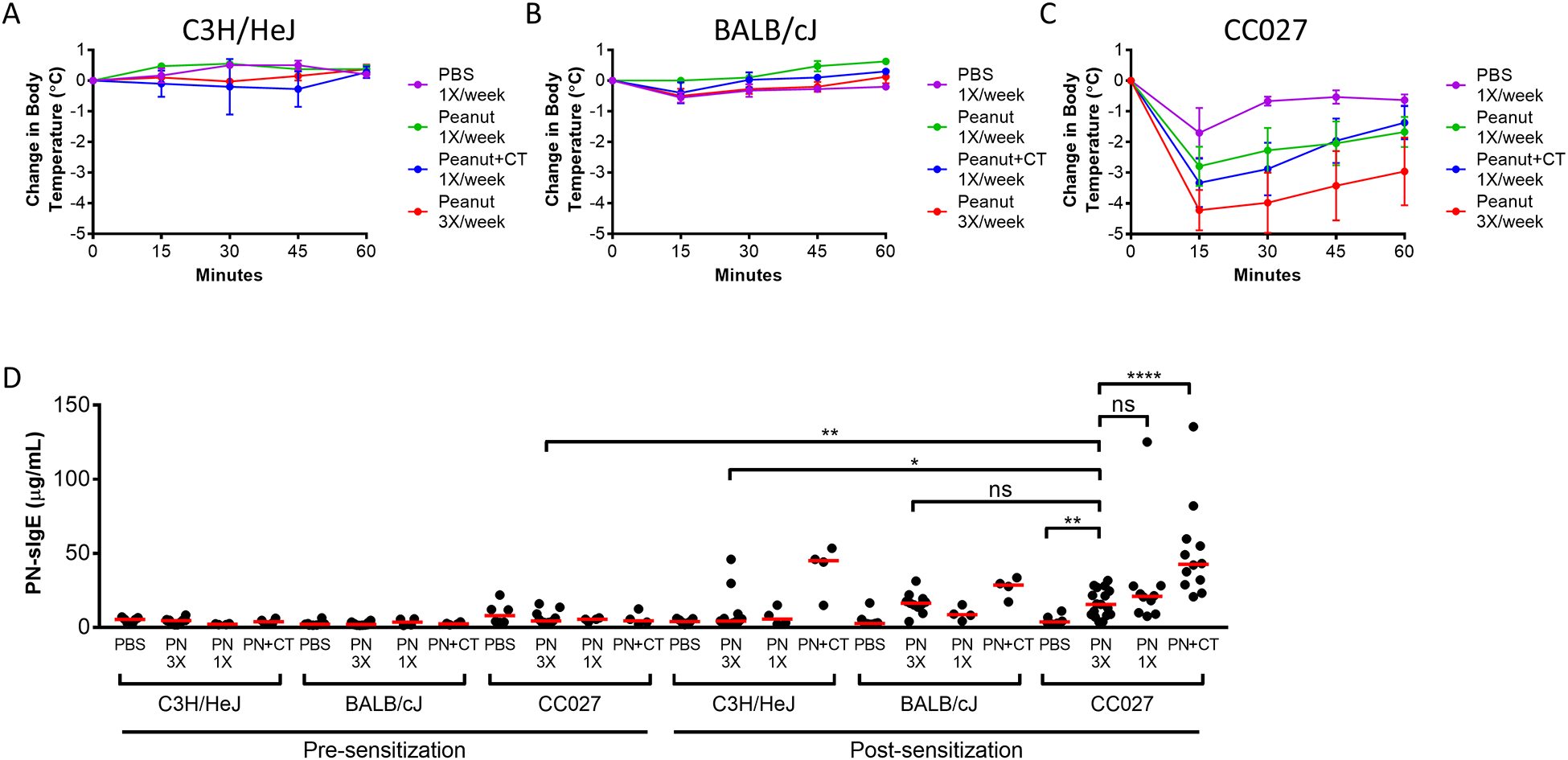
Anaphylaxis in peanut-sensitized mice following oral peanut challenge. Body temperatures after oral challenge with peanut extract in C3H/HeJ (A), BALB/cJ (B), and CC027/GeniUnc (C) mice sensitized with PBS, peanut extract once per week (PN 1X), peanut extract plus Cholera toxin once per week (PN+CT), or peanut extract three times per week (PN 3X). Peanut-specific IgE quantities in mice pre- and post-sensitization (D). Mann-Whitney U Test *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Discussion:
An accurate translation between small animal models and human health outcomes requires that models accurately recapitulate key aspects of the human disease. Previously, we utilized a mouse model of food allergy that requires IP challenge with peanut extract to elicit an anaphylactic response after sensitization with peanut and a Th2-skewing adjuvant (24, 27). However, a mouse that reacts on oral challenge would provide a more physiologically-relevant platform to study both the etiology of the disease as well as potential treatments. Within the human population, increasing evidence has shown that host genetic variation impacts allergic responses. A twin-study (19) estimated the heritability (proportion of genetic contribution) to peanut allergy at approximately 0.8. However, identification of genetic variants contributing to peanut allergy responses and outcomes has been limited to associations with the MHC locus and others associated with asthma and eczema (20, 39, 40). Undoubtedly, there are additional genetically variable factors driving propensity for, and severity of allergic responses to peanut. Therefore, we sought to determine whether genetic variation between mouse strains could explain variation in food allergy disease severity, and whether we could develop a more relevant oral challenge model by assessing genetically diverse inbred mouse strains.
Here, we described the use of 16 strains from the CC GRP to screen for an orally reacting animal model of peanut allergy. The CC strains are a set of reproducible inbred strains with high genetic diversity throughout the genome (34) and the CC has been used to both identify genetic factors driving aberrant disease outcomes (41–43), but also has enabled the development of more relevant models of human disease responses (14, 35, 44). We identified a single strain, CC027/GeniUnc as a promising model of food allergy. CC027/GeniUnc experiences a severe systemic reaction, evidenced by decreased body temperature following OFC with peanut extract, whereas the other 15 CC strains (as well as the well-studied inbred strains C3H/HeJ, C57BL/6J, and BALB/cJ) did not react accordingly. CC027/GeniUnc mice produce detectable levels of IL-4 protein and produce PNsIgE, Ara h 1-sIgE, Ara h 2-sIgE, Ara h 3-sIgE, similar to peanut allergy in humans (45). Quantities of IL-4 and IL-4Rα mRNA in the jejunum were similar among strains, although protein levels of IL-4 and IL-4Rα may be different. CC027/GeniUnc have increased levels of Th2-promoting transcription factor, Gata3 mRNA relative to C3H/HeJ mice, which do not react on oral challenge. Notably, our experimental design measured secreted cytokines and gene transcripts one week after oral challenge; it would be interesting to investigate changes in these levels at several time points before and after challenge in future studies to better understand the dynamic changes in activation and responses to allergen across different time points. Also, similar to human disease, PNsIgE does not correlate with disease severity in these mice. Furthermore, CC027/Geni/Unc have a lower number of Tregs based on flow cytometry data as well as lower levels of the important regulatory cytokine IL-10 at the protein and mRNA levels. Together, these results reveal a model of peanut allergy that, like other models (22), produce IL-4 in response to the allergen, a decreased regulatory response, but also demonstrates signs of a severe, systemic reaction on oral challenge with the allergen, making it a highly relevant model recapitulating key features of peanut allergy in humans.
As in human food allergy, the exact mechanistic causes of the increased reactivity of CC027/GeniUnc need to be further studied. Our findings suggest many potential contributing factors likely driven by the underlying genetic differences in these mice. As already stated, PNsIgE does not correlate with reaction severity, signifying that differences beyond IgE levels must be important for the severe oral reactions observed. In addition to hallmarks of acquired immune differences in CC027/GeniUnc, this strain has a greater quantity of basophils and mast cells; those effector cells responsible for the manifestations of allergic symptoms. Recent reports suggest an important interplay between activating and inhibitory signals from the surface of mast cells on allergic disease (46). While we did not assess mast cell activation, we found that the increased numbers of basophils possess less of the inhibitory receptor CD200R1 in CC027/GeniUnc than C3H/HeJ or C57BL/6J, similar to what has been reported for subjects with birch pollen allergy (47). Thus, CC027/GeniUnc may have a larger number of more easily activated effector cells than the other less-reactive strains. Although histamine levels are similar between strains, the possibility remains that CC027/GeniUnc may have a higher sensitivity to histamine, as has been shown previously in other mouse strains (48).
Furthermore, we demonstrated that CC027/GeniUnc absorbed higher levels of Ara h 2 protein into their blood stream during OFC than either C3H/HeJ or C57BL/6J, and these levels of serum Ara h 2 in CC027/GeniUnc correlated with reaction severity. Ara h 1 and Ara h 3 were not detectable in any strain, which could be due to the time frame we bled the mice following OFC since these proteins have lower stability compared to Ara h 2. C57BL/6J had detectable levels of serum Ara h 2 protein following OFC, but did not exhibit any signs of a systemic reaction or any detectable serum MMCP-1 following OFC. Taken together, these findings suggest mast cells in C57BL/6J are difficult to degranulate compared to CC027/GeniUnc mast cells. However, C3H/HeJ had high levels of MMCP-1 following OFC, but no serum Ara h 2 or symptoms of anaphylaxis. It is possible that only local mast cells in the mucosa degranulate in C3H/HeJ following oral challenge whereas CC027/GeniUnc experienced both local and systemic degranulation. The positive correlation observed between serum Ara h 2 levels and reaction severity in CC027/GeniUnc suggests that CC027/GeniUnc experience more severe reactions because of increased allergen absorption into their blood stream, which can trigger anaphylaxis by degranulation of mast cells, basophils, and/or neutrophils. These findings suggest that both Ara h 2 absorption into systemic circulation along with readily-degranulating effector cells is required for anaphylaxis upon OFC. Increased Ara h 2 absorption could be due to increased gut permeability in CC027/GeniUnc. Though a role for intestinal permeability in food allergy has been suggested (49, 50), attempts by our own group and others (51) to measure Ara h 2 in human serum following ingestion has proven difficult and inconclusive. Thus, CC027/GeniUnc offer insight into a potential disease mechanism that is currently difficult to investigate in humans. Future investigation of the uptake of Ara h 2 through the gastrointestinal tract is needed.
By also investigating whether CC027/GeniUnc mice could be sensitized with peanut extract alone, we have further validated the utility of this strain in food allergy research. The model in CC027/GeniUnc mice has addressed the two main weaknesses of current food allergy animal models: sensitization requiring an adjuvant such as Cholera toxin or Staphylococcal enterotoxin B, and anaphylactic reactions on IP challenge, not OFC. CC027/GeniUnc can be orally sensitized with peanut extract alone, and react orally to peanut, which more closely mimic our current understanding of the human disease. The ability of CC027/GeniUnc mice to be sensitized in the absence of Cholera toxin was further suggested by increased levels of peanut-specific IgE measured at baseline. Further studies are needed to understand this IgE production in the absence of a known antigen exposure in a subset of mice, and the underlying mechanisms leading to failure of oral tolerance in CC027/GeniUnc mice.
CC027/GeniUnc represents a highly relevant model of peanut allergy to the field at-large. This small-animal model should allow for more robust evaluation of therapeutic treatments in a pre-clinical setting prior to transition into clinical trials. Leading investigational treatments in the field include various routes of peanut immunotherapy including oral immunotherapy (OIT), sublingual immunotherapy (SLIT), and epicutaneous immunotherapy (EPIT). Despite promising results from OIT, SLIT, and EPIT studies (9, 10, 52, 53), these therapies have limitations including daily dosing, side effects, and difficulty in achieving long-term tolerance after stopping therapy. Therefore, new therapies that induce immunologic tolerance are needed. CC027/GeniUnc provides a pre-clinical model to develop these therapies and study the effects on the development of oral tolerance. Furthermore, genetic dissection of the repressive and enhancing phenotypes observed across mouse strains can lead to the identification of novel genes and pathways that may be critical in promoting peanut allergy within the human population. More broadly, our results highlight the utility of integrating the experimental robustness of inbred small animal models of disease with defined and broad genetic diversity in attempting to better understand and address human disease needs.
Supplementary Material
Key Messages:
Screen of 16 genetically diverse Collaborative Cross strains was used to identify one strain, CC027/GeniUnc, that reacts severely following oral sensitization and oral challenge with peanut extract.
The Collaborative Cross provides a platform to study the immunology as well as contributing factors such as genetics and environment to the development of food allergy.
Acknowledgements:
The authors would like to thank Soheila Maleki for providing purified Ara h 1, 2, and 3. The UNC Flow Cytometry Core Facility and Systems Genetics Core Facility are supported in part by P30 CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center.
Funding: SOM Office of Research and TraCS Translational Team Science Award (Grant # TTSA017P1, TTSA017P2) to AWB, FPMV, MDK and MTF; U19AI100625 to FPMV and MTF.
Abbreviations:
- OFC
Oral Food Challenge
- CC
Collaborative Cross
- GRP
Genetic Reference Panel
- DO
Diversity Outbred
- IP
Intraperitoneal
- OIT
Oral Immunotherapy
- SLIT
Sublingual Immunotherapy
- EPIT
Epicutaneous Immunotherapy
- PNsIgE
Peanut-specific IgE
- PNsIgG1
Peanut-specific IgG1
- PNsIgG2a/2c
Peanut-specific IgG2a/2c
- sIgE
specific IgE
- CT
Cholera toxin
Footnotes
Conflicts of Interest: The authors declare no conflicts of interest.
References:
- 1.Sicherer SH, Munoz-Furlong A, Godbold JH, Sampson HA. US prevalence of self-reported peanut, tree nut, and sesame allergy: 11-year follow-up. The Journal of allergy and clinical immunology. 2010;125(6):1322–6. [DOI] [PubMed] [Google Scholar]
- 2.Avery NJ, King RM, Knight S, Hourihane JO. Assessment of quality of life in children with peanut allergy. Pediatr Allergy Immunol. 2003;14(5):378–82. [DOI] [PubMed] [Google Scholar]
- 3.Sicherer SH, Sampson HA. Food allergy: Epidemiology, pathogenesis, diagnosis, and treatment. J Allergy Clin Immunol. 2014;133(2):291–307; quiz 8. [DOI] [PubMed] [Google Scholar]
- 4.Iweala OI, Burks AW. Food Allergy: Our Evolving Understanding of Its Pathogenesis, Prevention, and Treatment. Current allergy and asthma reports. 2016;16(5):37. [DOI] [PubMed] [Google Scholar]
- 5.Jones SM, Pons L, Roberts JL, Scurlock AM, Perry TT, Kulis M, et al. Clinical efficacy and immune regulation with peanut oral immunotherapy. The Journal of allergy and clinical immunology. 2009;124(2):292–300, e1–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varshney P, Jones SM, Scurlock AM, Perry TT, Kemper A, Steele P, et al. A randomized controlled study of peanut oral immunotherapy: clinical desensitization and modulation of the allergic response. J Allergy Clin Immunol. 2011;127(3):654–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones SM, Sicherer SH, Burks AW, Leung DY, Lindblad RW, Dawson P, et al. Epicutaneous immunotherapy for the treatment of peanut allergy in children and young adults. The Journal of allergy and clinical immunology. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burks AW, Wood RA, Jones SM, Sicherer SH, Fleischer DM, Scurlock AM, et al. Sublingual immunotherapy for peanut allergy: Long-term follow-up of a randomized multicenter trial. The Journal of allergy and clinical immunology. 2015;135(5):1240–8. e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim EH, Bird JA, Kulis M, Laubach S, Pons L, Shreffler W, et al. Sublingual immunotherapy for peanut allergy: clinical and immunologic evidence of desensitization. J Allergy Clin Immunol. 2011;127(3):640–6 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleischer DM, Burks AW, Vickery BP, Scurlock AM, Wood RA, Jones SM, et al. Sublingual immunotherapy for peanut allergy: a randomized, double-blind, placebo-controlled multicenter trial. J Allergy Clin Immunol. 2013;131(1):119–27 e1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sampath V, Tupa D, Graham MT, Chatila TA, Spergel JM, Nadeau KC. Deciphering the black box of food allergy mechanisms. Ann Allergy Asthma Immunol. 2017;118(1):21–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu W, Freeland DMH, Nadeau KC. Food allergy: immune mechanisms, diagnosis and immunotherapy. Nature reviews Immunology. 2016;16(12):751–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warren HS, Tompkins RG, Moldawer LL, Seok J, Xu W, Mindrinos MN, et al. Mice are not men. Proc Natl Acad Sci U S A. 2015;112(4):E345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rasmussen AL, Okumura A, Ferris MT, Green R, Feldmann F, Kelly SM, et al. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science (New York, NY). 2014;346(6212):987–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cichocki JA, Furuya S, Venkatratnam A, McDonald TJ, Knap AH, Wade T, et al. Characterization of Variability in Toxicokinetics and Toxicodynamics of Tetrachloroethylene Using the Collaborative Cross Mouse Population. Environmental health perspectives. 2017;125(5):057006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andreux PA, Williams EG, Koutnikova H, Houtkooper RH, Champy MF, Henry H, et al. Systems genetics of metabolism: the use of the BXD murine reference panel for multiscalar integration of traits. Cell. 2012;150(6):1287–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics. 2012;190(2):389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Churchill GA, Gatti DM, Munger SC, Svenson KL. The Diversity Outbred mouse population. Mammalian genome : official journal of the International Mammalian Genome Society. 2012;23(9–10):713–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sicherer SH, Furlong TJ, Maes HH, Desnick RJ, Sampson HA, Gelb BD. Genetics of peanut allergy: a twin study. The Journal of allergy and clinical immunology. 2000;106(1 Pt 1):53–6. [DOI] [PubMed] [Google Scholar]
- 20.Hong X, Hao K, Ladd-Acosta C, Hansen KD, Tsai HJ, Liu X, et al. Genome-wide association study identifies peanut allergy-specific loci and evidence of epigenetic mediation in US children. Nature communications. 2015;6:6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen-Jarolim E, Pali-Scholl I, Roth-Walter F. Outstanding animal studies in allergy I. From asthma to food allergy and anaphylaxis. Current opinion in allergy and clinical immunology. 2017;17(3):169–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tordesillas L, Goswami R, Benede S, Grishina G, Dunkin D, Jarvinen KM, et al. Skin exposure promotes a Th2-dependent sensitization to peanut allergens. J Clin Invest. 2014;124(11):4965–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li XM, Serebrisky D, Lee SY, Huang CK, Bardina L, Schofield BH, et al. A murine model of peanut anaphylaxis: T- and B-cell responses to a major peanut allergen mimic human responses. The Journal of allergy and clinical immunology. 2000;106(1 Pt 1):150–8. [DOI] [PubMed] [Google Scholar]
- 24.Kulis M, Chen X, Lew J, Wang Q, Patel OP, Zhuang Y, et al. The 2S albumin allergens of Arachis hypogaea, Ara h 2 and Ara h 6, are the major elicitors of anaphylaxis and can effectively desensitize peanut-allergic mice. Clin Exp Allergy. 2012;42(2):326–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganeshan K, Neilsen CV, Hadsaitong A, Schleimer RP, Luo X, Bryce PJ. Impairing oral tolerance promotes allergy and anaphylaxis: a new murine food allergy model. J Allergy Clin Immunol. 2009;123(1):231–8 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulis M, Gorentla B, Burks AW, Zhong XP. Type B CpG oligodeoxynucleotides induce Th1 responses to peanut antigens: modulation of sensitization and utility in a truncated immunotherapy regimen in mice. Mol Nutr Food Res. 2013;57(5):906–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orgel KA, Duan S, Wright BL, Maleki SJ, Wolf JC, Vickery BP, et al. Exploiting CD22 on antigen-specific B cells to prevent allergy to the major peanut allergen Ara h 2. J Allergy Clin Immunol. 2017;139(1):366–9 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burton OT, Stranks AJ, Tamayo JM, Koleoglou KJ, Schwartz LB, Oettgen HC. A humanized mouse model of anaphylactic peanut allergy. J Allergy Clin Immunol. 2017;139(1):314–22 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berin MC, Mayer L. Immunophysiology of experimental food allergy. Mucosal immunology. 2009;2(1):24–32. [DOI] [PubMed] [Google Scholar]
- 30.Bryce PJ, Falahati R, Kenney LL, Leung J, Bebbington C, Tomasevic N, et al. Humanized mouse model of mast cell-mediated passive cutaneous anaphylaxis and passive systemic anaphylaxis. J Allergy Clin Immunol. 2016;138(3):769–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun J, Arias K, Alvarez D, Fattouh R, Walker T, Goncharova S, et al. Impact of CD40 ligand, B cells, and mast cells in peanut-induced anaphylactic responses. J Immunol. 2007;179(10):6696–703. [DOI] [PubMed] [Google Scholar]
- 32.Welsh CE, Miller DR, Manly KF, Wang J, McMillan L, Morahan G, et al. Status and access to the Collaborative Cross population. Mammalian genome : official journal of the International Mammalian Genome Society. 2012;23(9–10):706–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H, Hu Q, D’Ercole AJ, Ye P. Histone deacetylase 11 regulates oligodendrocyte-specific gene expression and cell development in OL-1 oligodendroglia cells. Glia. 2009;57(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srivastava A, Morgan AP, Najarian ML, Sarsani VK, Sigmon JS, Shorter JR, et al. Genomes of the Mouse Collaborative Cross. Genetics. 2017;206(2):537–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogala AR, Morgan AP, Christensen AM, Gooch TJ, Bell TA, Miller DR, et al. The Collaborative Cross as a resource for modeling human disease: CC011/Unc, a new mouse model for spontaneous colitis. Mammalian genome : official journal of the International Mammalian Genome Society. 2014;25(3–4):95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blazquez AB, Berin MC. Gastrointestinal dendritic cells promote Th2 skewing via OX40L. J Immunol. 2008;180(7):4441–50. [DOI] [PubMed] [Google Scholar]
- 37.Stone KD, Prussin C, Metcalfe DD. IgE, mast cells, basophils, and eosinophils. The Journal of allergy and clinical immunology. 2010;125(2 Suppl 2):S73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barclay AN, Wright GJ, Brooke G, Brown MH. CD200 and membrane protein interactions in the control of myeloid cells. Trends in immunology. 2002;23(6):285–90. [DOI] [PubMed] [Google Scholar]
- 39.Shreffler WG, Charlop-Powers Z, Sicherer SH. Lack of association of HLA class II alleles with peanut allergy. Ann Allergy Asthma Immunol. 2006;96(6):865–9. [DOI] [PubMed] [Google Scholar]
- 40.Asai Y, Eslami A, van Ginkel CD, Akhabir L, Wan M, Ellis G, et al. Genome-wide association study and meta-analysis in multiple populations identifies new loci for peanut allergy and establishes c11orf30/EMSY as a genetic risk factor for food allergy. The Journal of allergy and clinical immunology. 2017. [DOI] [PubMed] [Google Scholar]
- 41.Gralinski LE, Menachery VD, Morgan AP, Totura AL, Beall A, Kocher J, et al. Allelic Variation in the Toll-Like Receptor Adaptor Protein Ticam2 Contributes to SARS-Coronavirus Pathogenesis in Mice. G3 (Bethesda, Md). 2017;7(6):1653–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Durrant C, Tayem H, Yalcin B, Cleak J, Goodstadt L, de Villena FP, et al. Collaborative Cross mice and their power to map host susceptibility to Aspergillus fumigatus infection. Genome Res. 2011;21(8):1239–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelada SN, Carpenter DE, Aylor DL, Chines P, Rutledge H, Chesler EJ, et al. Integrative genetic analysis of allergic inflammation in the murine lung. American journal of respiratory cell and molecular biology. 2014;51(3):436–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graham JB, Swarts JL, Wilkins C, Thomas S, Green R, Sekine A, et al. A Mouse Model of Chronic West Nile Virus Disease. PLoS pathogens. 2016;12(11):e1005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turcanu V, Winterbotham M, Kelleher P, Lack G. Peanut-specific B and T cell responses are correlated in peanut-allergic but not in non-allergic individuals. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2008;38(7):1132–9. [DOI] [PubMed] [Google Scholar]
- 46.Gibbs BF, Sabato V, Bridts CH, Ebo DG, Ben-Zimra M, Levi-Schaffer F. Expressions and inhibitory functions of CD300a receptors on purified human basophils. Experimental dermatology. 2012;21(11):884–6. [DOI] [PubMed] [Google Scholar]
- 47.Burton OT, Logsdon SL, Zhou JS, Medina-Tamayo J, Abdel-Gadir A, Noval Rivas M, et al. Oral immunotherapy induces IgG antibodies that act through FcgammaRIIb to suppress IgE-mediated hypersensitivity. The Journal of allergy and clinical immunology. 2014;134(6):1310–7.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arumugam M, Ahrens R, Osterfeld H, Kottyan LC, Shang X, Maclennan JA, et al. Increased susceptibility of 129SvEvBrd mice to IgE-Mast cell mediated anaphylaxis. BMC Immunol. 2011;12:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perrier C, Corthesy B. Gut permeability and food allergies. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2011;41(1):20–8. [DOI] [PubMed] [Google Scholar]
- 50.Ventura MT, Polimeno L, Amoruso AC, Gatti F, Annoscia E, Marinaro M, et al. Intestinal permeability in patients with adverse reactions to food. Digestive and liver disease : official journal of the Italian Society of Gastroenterology and the Italian Association for the Study of the Liver. 2006;38(10):732–6. [DOI] [PubMed] [Google Scholar]
- 51.JanssenDuijghuijsen LM, Wichers HJ, van Norren K, Keijer J, Baumert JL, de Jong GA, et al. Detection of peanut allergen in human blood after consumption of peanuts is skewed by endogenous immunoglobulins. J Immunol Methods. 2017;440:52–7. [DOI] [PubMed] [Google Scholar]
- 52.Vickery BP, Berglund JP, Burk CM, Fine JP, Kim EH, Kim JI, et al. Early oral immunotherapy in peanut-allergic preschool children is safe and highly effective. The Journal of allergy and clinical immunology. 2017;139(1):173–81.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones SM, Sicherer SH, Burks AW, Leung DY, Lindblad RW, Dawson P, et al. Epicutaneous immunotherapy for the treatment of peanut allergy in children and young adults. The Journal of allergy and clinical immunology. 2017;139(4):1242–52.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


