Abstract
Wilms tumor (WT) is the most common malignant renal neoplasm in children; however, the underlying molecular mechanisms are not well understood. According to the competing endogenous RNA (ceRNA) theory, long non-coding RNAs (lncRNAs) can regulate the expression of target genes by adsorbing microRNAs (miRNAs/miRs). However, the role of lncRNAs in WT has not been fully elucidated. The aim of the present study was to construct a ceRNA network to identify the potential lncRNAs involved in WT. The expression profiles of lncRNAs, miRNAs and mRNAs in 120 WT and six normal tissues were obtained from the Therapeutically Applicable Research to Generate Effective Treatments database. A total of 442 lncRNAs, 214 miRNAs and 4,912 mRNAs were identified as differentially expressed in WT and were enriched in 472 Gene Ontology terms (355 biological processes, 89 cellular components and 29 molecular functions) and 18 Kyoto Encyclopedia of Genes and Genomes pathways. A lncRNA-miRNA-mRNA ceRNA network of WT consisting of with 32 lncRNAs, 14 miRNAs and 158 mRNAs was constructed, based on the bioinformatics analysis of the miR target prediction database and the miRNAcode, miRTarBase and TargetScan databases. Subsequently, three lncRNAs, three miRNAs and 17 mRNAs, which had a significant effect on the overall survival rate of patients with WT, were identified based on the survival analysis. The three lncRNAs were also differentially expressed in the late and early stages of WT and were validated using the GSE66405 dataset obtained from the Gene Expression Omnibus database. In conclusion, the present study generated a specific lncRNA-related ceRNA network of WT, which may provide a novel perspective on the molecular mechanisms underlying the progression and prognosis of the disease.
Keywords: Wilms tumor, competitive endogenous RNA, long non-coding RNA, Therapeutically Applicable Research to Generate Effective Treatments database, prognosis
Introduction
Renal tumors are the second most common abdominal tumor in infants and children. Different types of renal tumor have distinct histological features, and Wilms tumor (WT) is the most frequently occurring type, accounting for ~95% of pediatric renal tumors in the United States (1). A total of seven in one million American children <15 years of age are affected by WT annually (2). In the past few decades, the prognosis of patients with WT has significantly improved. According to a statistic published in 2018, the overall 5-year survival rate is >90% (3). Nevertheless, the main therapeutic regimen for WT is still surgery combined with radiotherapy and chemotherapy (4). However, children who receive high-intensity radiotherapy and chemotherapy are more likely to experience complications, including renal impairment, cardiotoxicity, hepatotoxicity, delayed growth, infertility and secondary malignancies (5–9). Surgical resection also has a number of limitations, such as bilateral tumors, tumor in a solitary kidney and extensive pulmonary metastases (10). Therefore, understanding the molecular mechanisms underlying the progression of WT and developing molecular-targeted therapies are required.
Only 2% of the human genome comprises protein-coding genes (11), while the majority is transcribed into non-coding RNAs, including long non-coding RNAs (lncRNAs) and microRNAs (miRNAs/miRs) (12). lncRNAs range in length from 200 nucleotides to 100 kb (13). An increasing number of studies have reported that lncRNAs are involved in the biological behaviors of tumor cells, including proliferation, apoptosis, the cell cycle, invasion and migration (14–18). According to the theory of competitive endogenous RNA (ceRNA), lncRNAs act as a sponge for miRNAs, weakening the effects of miRNAs on mRNAs (19). Several other studies have confirmed this hypothesis and constructed ceRNA networks for various types of cancer (20–22). However, there is currently limited research on lncRNA-mediated ceRNAs and the role of lncRNAs in the prognosis of patients with WT (23–26). Therefore, the construction of a ceRNA network could aid in the discovery of novel molecular targets and the development of therapeutic strategies to improve the prognosis of patients with WT.
The aim of the present study was to construct a ceRNA network to identify potential lncRNAs involved in WT. The expression profiles of lncRNAs, miRNAs and mRNAs were obtained from the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) database. Furthermore, a lncRNA-miRNA-mRNA ceRNA network of WT was constructed by integrated analysis. Additionally, the prognostic value of lncRNAs in the ceRNA network was analyzed.
Materials and methods
Data collection and processing
RNA sequencing (RNA-Seq) data of 131 samples, miRNA sequencing (miRNA-Seq) data of 138 samples and clinical prognostic information of 655 samples were retrieved from the TARGET database via the Genomic Data Commons Data Portal (portal.gdc.cancer.gov). The inclusion criteria were as follows: i) Tissue samples with RNA-Seq data, miRNA-Seq data and clinical prognostic information; and ii) samples from primary tumors. A total of 120 primary tumor tissues and six adjacent normal kidney tissues were included in the present study. Perl (version 5.28.0; http://www.perl.org/) and R software (version 3.6.0; http://www.r-project.org/) were used for data processing. Differences in the expression of genes between the two groups were analyzed using the principal component analysis (PCA) tool (version 1.2.0; http://github.com/kevinblighe/PCAtools) for R. Data used for validating the results were downloaded from the Gene Expression Omnibus (GEO) dataset GSE66405 (27), which consisted of 28 tumor and four adjacent normal kidney tissues. A flowchart illustrating the generation of the ceRNA network is presented in Fig. S1.
Identification of differentially expressed genes (DEGs)
After data normalization, the ‘edgeR’ package (version 3.24.3) for R (28), with the limma method (version 3.38.3) (29), were used to identify the differentially expressed lncRNAs (DElncRNAs), mRNAs (DEmRNAs) and miRNAs (DEmiRNAs) between the WT and normal tissues. The cut-off value for DEGs was |log2 fold-change (FC)|>1 with a false discovery rate (FDR) of <0.05. The DElncRNAs, DEmRNAs and DEmiRNAs were visualized by volcano maps and heat maps using the ‘ggplot2’ (version 3.2.0; http://ggplot2.tidyverse.org/) and ‘pheatmap’ (version 1.10.1; https://cran.r-project.org/web/ packages /pheatmap/index.html) R packages.
Functional and pathway enrichment analyses
To further understand the molecular mechanisms of WT, the ‘clusterProfiler’ (version 3.10.1) R package (30) was used to analyze the Gene Ontology (GO) annotation (http://geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment (https://www.kegg.jp/) of DEmRNAs in the ceRNA network. A corrected P-value of <0.01 was considered to indicate a statistically significant difference. The results are presented as bubble plots, generated using the ‘ggplot2’ package in R.
Construction of the ceRNA network
The miRcode (version 11; www.mircode.org) database was used to predict interactions between DElncRNAs and DEmiRNAs. Then, the miRNA target prediction database miRDB (version 6.0; www.mirdb.org) and TargetScan (release 7.2; www.targetscan.org) and miRTarBase (release 7.0; mirtarbase.mbc.nctu.edu.tw) databases were used to predict the target mRNAs of the DEmiRNAs. Only mRNAs present in all three databases were considered as candidate mRNAs that interacted with DEmRNAs. The overlapping mRNAs were considered to be target DEmRNAs of the DEmiRNAs. The analysis was carried out using Perl. Finally, the DElncRNA-DEmiRNA-DEmRNA network was constructed and visualized using Cytoscape software (version 3.6.1; http://cytoscape.org/).
Survival analysis
The ‘survival’ package in R was used to analyze DElncRNAs, DEmiRNAs and DEmRNAs associated with the prognosis of WT. All samples were divided into high or low expression groups, according to the median RNA expression level. The Kaplan-Meier method and the log-rank test were used to generate and compare survival curves. P<0.05 was considered to indicate a statistically significant difference. Furthermore, Pearson correlation analyses by R software (version 3.6.0; http://www.r-project.org/) were performed to examine the correlation between the expression of the DElncRNAs and the corresponding DEmRNAs in the ceRNA network.
Validation of DElncRNAs
The DElncRNAs were compared according to the National Wilms Tumor Study staging system (4). In addition, the expression profiles of the DElncRNAs were extracted from the GSE66405 gene expression matrix (27). The DElncRNA expression profiles in the tumor and normal groups were compared using an unpaired Student's t-test. Statistical analysis was performed using GraphPad Prism software (version 8.0; GraphPad Software, Inc.).
Results
Identification of the DElncRNAs, DEmiRNAs and DEmRNAs
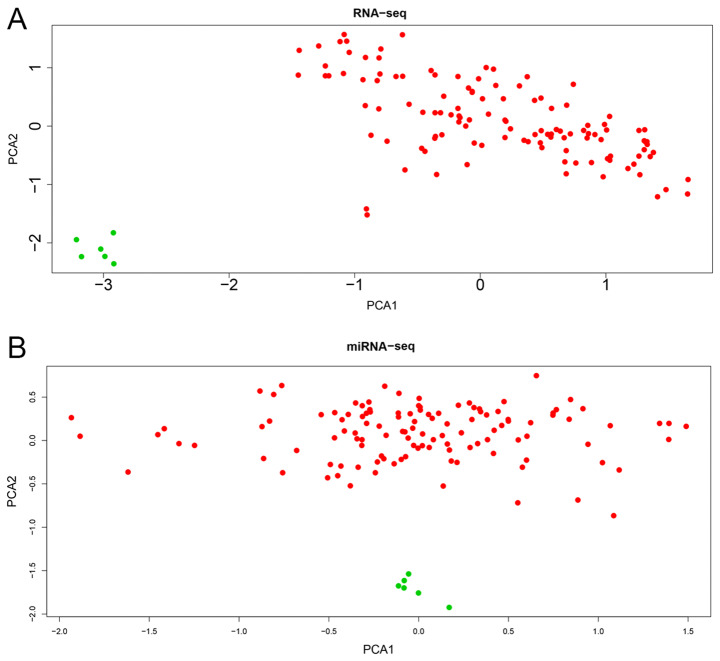
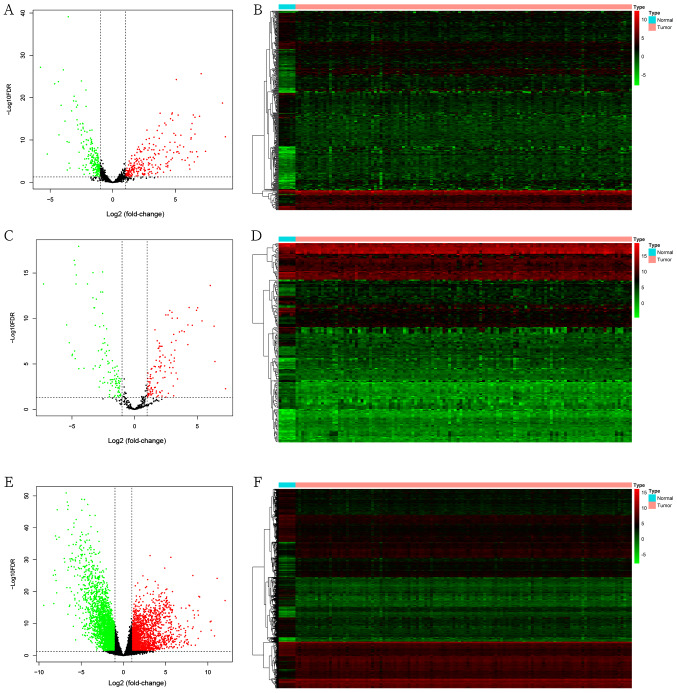
The present study included 120 primary tumor tissues and six adjacent normal kidney tissues. The clinical features of the patients are presented in Table I. The RNA-seq (mRNAs and lncRNAs) and miRNAs in the two groups were expressed in two clear groups in the PCA plot (Fig. 1). Subsequently, 442 DElncRNAs (234 upregulated and 208 downregulated; Fig. 2A and B), 214 DEmiRNAs (120 upregulated and 94 downregulated; Fig. 2C and D) and 4,912 DEmRNAs (2056 upregulated and 2856 downregulated; Fig. 2E and F) between the WT and normal tissues were identified according to the cut-off criteria (|log2FC|>1; FDR <0.05).
Table I.
Clinical characteristics of patients with Wilms tumor in the Therapeutically Applicable Research to Generate Effective Treatments database.
| Characteristic | Number (%) |
|---|---|
| Sex | |
| Male | 53 (44.2) |
| Female | 67 (55.8) |
| Ethnicity | |
| Black | 19 (15.8) |
| White | 89 (74.2) |
| Other | 5 (4.2) |
| Not reported | 7 (5.8) |
| Age at diagnosis | |
| <24 months | 16 (13.3) |
| >24 months | 104 (86.7) |
| Histology | |
| DAWT | 40 (33.3) |
| FHWT | 80 (66.7) |
| NMTS stage | |
| I | 15 (12.5) |
| II | 48 (40.0) |
| III | 44 (36.7) |
| IV | 12 (10.0) |
| V | 1 (0.8) |
| Progression or relapse | |
| Yes | 95 (79.2) |
| No | 25 (20.8) |
FHWT, favorable histology Wilms tumor; DAWT, diffuse anaplastic histology Wilms tumor; NMTS, National Wilms Tumor Study.
Figure 1.
PCA plots of RNA-seq and miRNA-seq expression. (A) PCA plot of >60,000 RNA-seq expression, including lncRNAs and mRNAs. (B) PCA plot of >2,000 miRNA-seq expression. The red and green dots represent tumor tissues and normal tissues, respectively. PCA, principal component analysis; seq, sequencing; miRNA, microRNA.
Figure 2.
Differential expression of lncRNAs, miRNAs and mRNAs. (A) Volcano plot and (B) heatmap of DElncRNAs. (C) Volcano plot and (D) heatmap of DEmiRNAs. (E) Volcano plot and (F) heatmap of DEmRNAs. In the volcano plots and heatmaps, red represents upregulated genes, green represents downregulated genes and black represents unchanged genes. A total of 442 DElncRNAs (234 upregulated and 208 downregulated), 214 DEmiRNAs (120 upregulated and 94 downregulated) and 4,912 DEmRNAs (2,056 upregulated and 2,856 downregulated) were identified according to the cut-off criteria (|log2 fold-change|>1 and FDR <0.05). DE, differentially expressed; lncRNA, long non-coding RNA; miRNA, microRNA; FDR, false discovery rate.
Functional and pathway enrichment analyses
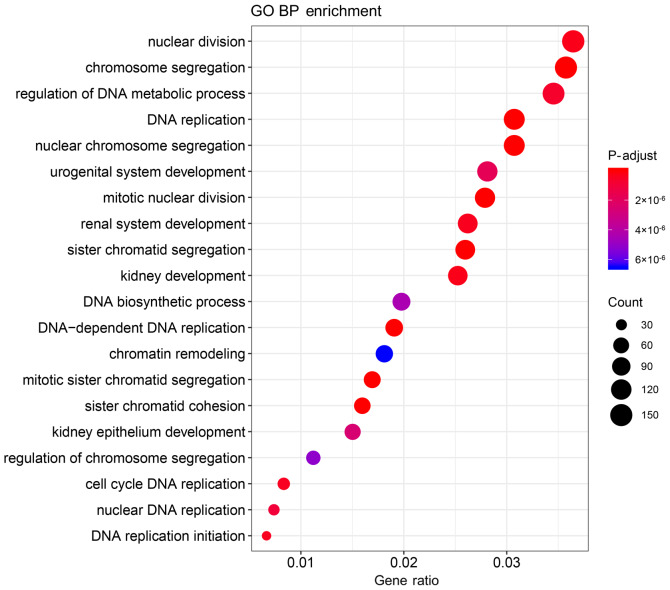
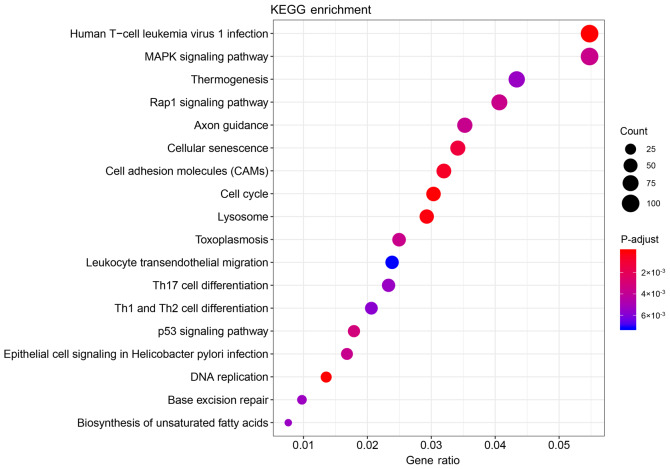
The functions and pathways of the 4,912 DEmRNAs were investigated using the ‘clusterProfiler’ R package. The results suggested that 473 GO terms (355 biological processes, 89 cell components and 29 molecular functions) and 18 KEGG pathways were significantly enriched in the DEmRNAs (Tables SI and SII). The top 20 significantly enriched biological processes of the GO terms are presented in Table II and Fig. 3, and the significantly enriched KEGG pathways are presented in Table III and Fig. 4.
Table II.
Top 20 enriched biological processes of GO analysis in Wilms tumor.
| ID | GO term | Count | P-value |
|---|---|---|---|
| GO:0006261 | DNA-dependent DNA replication | 80 | 3.73×10−13 |
| GO:0006260 | DNA replication | 129 | 1.55×10−12 |
| GO:0007059 | Chromosome segregation | 150 | 2.25×10−12 |
| GO:0000819 | Sister chromatid segregation | 109 | 2.25×10−12 |
| GO:0140014 | Mitotic nuclear division | 117 | 2.52×10−10 |
| GO:0098813 | Nuclear chromosome segregation | 129 | 3.17×10−10 |
| GO:0007062 | Sister chromatid cohesion | 67 | 3.99×10−9 |
| GO:0000070 | Mitotic sister chromatid segregation | 71 | 7.23×10−9 |
| GO:0000280 | Nuclear division | 153 | 3.08×10−7 |
| GO:0001822 | Kidney development | 106 | 3.08×10−7 |
| GO:0072001 | Renal system development | 110 | 4.20×10−7 |
| GO:0006270 | DNA replication initiation | 28 | 4.20×10−7 |
| GO:0044786 | Cell cycle DNA replication | 35 | 4.73×10−7 |
| GO:0051052 | Regulation of DNA metabolic process | 145 | 7.00×10−7 |
| GO:0033260 | Nuclear DNA replication | 31 | 1.02×10−6 |
| GO:0001655 | Urogenital system development | 118 | 1.78×10−6 |
| GO:0072073 | Kidney epithelium development | 63 | 2.58×10−6 |
| GO:0071897 | DNA biosynthetic process | 83 | 4.43×10−6 |
| GO:0051983 | Regulation of chromosome segregation | 47 | 5.12×10−6 |
| GO:0006338 | Chromatin remodeling | 76 | 6.51×10−6 |
GO, Gene Ontology.
Figure 3.
Top 20 significantly enriched BPs of the GO analysis of Wilms tumor-related genes. BP, biological processes; GO, Gene Ontology.
Table III.
Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis of differentially expressed genes in Wilms tumor.
| ID | Description | Count | P-value |
|---|---|---|---|
| hsa03030 | DNA replication | 25 | 5.74×10−6 |
| hsa05166 | Human T-cell leukemia virus 1 infection | 101 | 9.80×10−6 |
| hsa04110 | Cell cycle | 56 | 4.65×10−5 |
| hsa04142 | Lysosome | 54 | 1.65×10−4 |
| hsa04514 | Cell adhesion molecules (CAMs) | 59 | 6.57×10−4 |
| hsa04218 | Cellular senescence | 63 | 1.25×10−3 |
| hsa04115 | p53 signaling pathway | 33 | 3.19×10−3 |
| hsa04015 | Rap1 signaling pathway | 75 | 3.63×10−3 |
| hsa04010 | MAPK signaling pathway | 101 | 3.63×10−3 |
| hsa05145 | Toxoplasmosis | 46 | 3.63×10−3 |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | 31 | 3.74×10−3 |
| hsa04360 | Axon guidance | 65 | 3.74×10−3 |
| hsa03410 | Base excision repair | 18 | 5.35×10−3 |
| hsa01040 | Biosynthesis of unsaturated fatty acids | 14 | 5.40×10−3 |
| hsa04659 | Th17 cell differentiation | 43 | 5.41×10−3 |
| hsa04714 | Thermogenesis | 80 | 5.41×10−3 |
| hsa04658 | Th1 and Th2 cell differentiation | 38 | 5.77×10−3 |
| hsa04670 | Leukocyte transendothelial migration | 44 | 7.20×10−3 |
Figure 4.
Significantly enriched KEGG pathways in Wilms tumor-related genes. KEGG, Kyoto Encyclopedia of Genes and Genomes.
Construction of the ceRNA network
Using the miRcode database, 206 miRNAs were identified as the targets of 45 DElncRNAs. The 206 miRNAs were then intersected with the DEmiRNAs to identify 16 miRNAs that were regulated in an opposing manner to the 33 corresponding DElncRNAs.
Subsequently, 158 target genes of the 14 miRNAs were predicted using the miRDB, TargetScan and miRTarBase databases
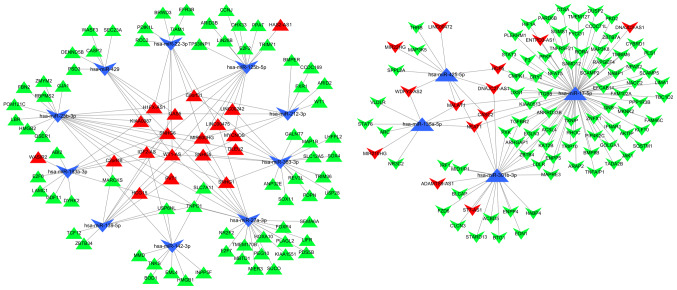
These 158 target genes were identified as DEmRNAs. Thus, 32 DElncRNAs, 14 DEmiRNAs and 158 DEmRNAs were included in the ceRNA network generated by Cytoscape software (Fig. 5).
Figure 5.
ceRNA network of the lncRNA-miRNA-mRNA interactions in Wilms tumor. Regular triangles and inverted triangles represent upregulated and downregulated genes, respectively. Red nodes represent lncRNAs, blue nodes represent miRNAs and green nodes represent mRNAs. ceRNA, competing endogenous RNA; lncRNA, long non-coding RNA; miRNA, microRNA; WT, Wilms tumor.
Survival analysis
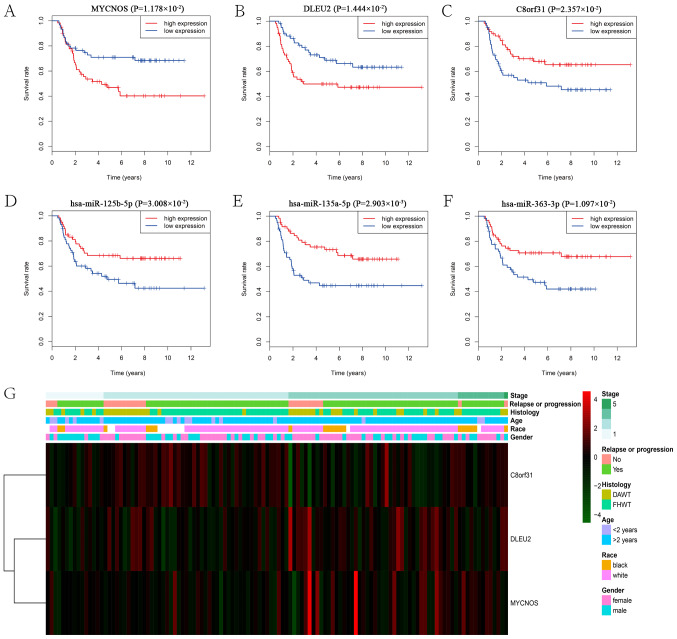
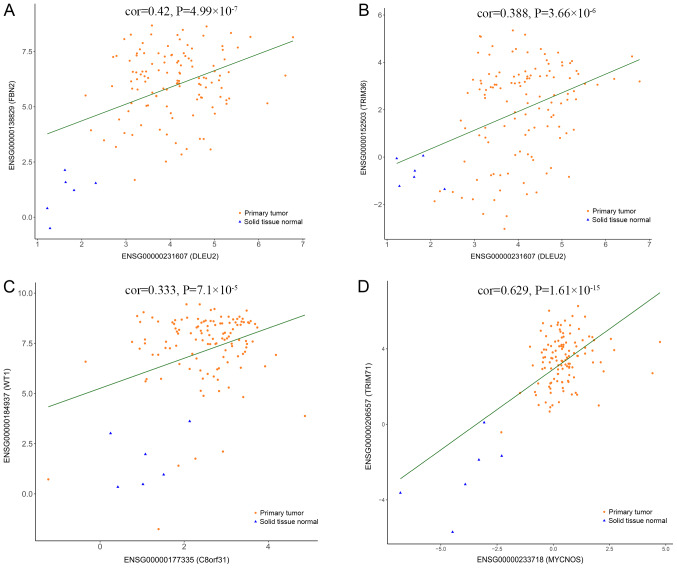
To further understand the effect of DElncRNAs, DEmiRNAs and DEmRNAs on the prognosis of WT, Kaplan-Meier curves of the 32 DElncRNAs, 14 DEmiRNAs and 158 DEmRNAs were analyzed for overall survival rate (OS). The results suggested that three DElncRNAs, three DEmiRNAs and 17 DEmRNAs were significantly associated with the OS (P<0.05). Of the three DElncRNAs, MYCN opposite strand (MYCNOS) and deleted in lymphocytic leukemia 2 (DLEU2) were negatively associated with the OS (P=1.178×10−2, P=1.444×10−2), while chromosome 8 open reading frame 31 (C8orf31) was positively associated (P=2.357×10−2) with the OS (Fig. 6A-C). All three of the DEmiRNAs (hsa-miR-135a-5p, hsa-miR-363-3p and hsa-miR-125b-5p) were positively associated (P=2.903×10−3, P=1.097×10−2, P=3.008×10−2, respectively) with the OS (Fig. 6D-F). The expression profile of the three DElncRNAs and the clinical features of the 120 patients with WT are presented in Fig. 6G. Among the 17 significant DEmRNAs (data not presented), five mRNAs [family with sequence similarity 102 member A (FAM102A), forkhead box P4 (FOXP4), WT1 transcription factor (WT1), tripartite motif (TRIM) containing TRIM36 and TRIM71] were negatively associated (P=2.453×10−2, P=4.4968×10−2, P=3.671×10−2, P=4.271×10−2, P=3.403×10−2) with the OS, whereas the other 12 mRNAs [fibrillin 2 (FBN2), glucosamine (N-acetyl)-6-sulfatase (GNS), Kruppel like factor 10 (KLF10), LIM domain and actin binding 1 (LIMA1), protein phosphatase 1 regulatory subunit 3B (PPP1R3B), pleckstrin and Sec7 domain containing 3 (PSD3), STAT3, STAT6, transmembrane protein (TMEM)127, UDP-glucuronate decarboxylase 1 (UXS1), very low density lipoprotein receptor (VLDLR) and zinc finger and BTB domain containing 4 (ZBTB4) were positively associated (P=2.287×10−2, P=3.143×10−2, P=1.635×10−2, P=4.366×10−3, P=3.102×10−2, P=1.148×10−2, P=4.057×10−2, P=3.406×10−2, P=2.462×10−2, P=2.985×10−2, P=4.977×10−2, P=4.012×10−2) with the OS. Moreover, a correlation analysis was performed between the three DElncRNAs and the 17 DEmRNAs associated with OS. The significantly correlated DElncRNAs and DEmRNAs are presented in Fig. 7. Considering that the three DElncRNAs displayed an association with the survival of patients with WT, univariate and multivariate Cox regression analyses of OS were performed with the three DElncRNAs and the clinical features (Table IV). The univariate analysis suggested that ethnicity, stage and MYCNOS expression significantly affected the OS of patients with WT (P<0.05; Table IV). The multivariate analysis suggested that stage and MYCNOS expression were independent prognostic indicators in patients with WT (P<0.05; Table IV).
Figure 6.
Survival analysis and expression profiles of the three DElncRNAs. Kaplan-Meier survival curves of the three DElncRNAs: (A) MYCNOS, (B) DLEU2 and (C) C8orf31, and the three DEmiRNAs: (D) hsa-miR-125b-5p, (E) hsa-miR-135a-5p and (F) hsa-miR-363-3p, associated with overall survival in WT. (G) Expression profile of the three DElncRNAs and the clinical features of the 120 patients with WT. DE, differentially expressed; lncRNA, long non-coding RNA; MYCNOS, MYCN opposite strand; DLEU2, deleted in lymphocytic leukemia 2; C8orf31, chromosome 8 open reading frame 31; miRNA, microRNA; miR, microRNA; WT, Wilms tumor; DAWT, diffuse anaplastic histology WT; FHWT, favorable histology WT.
Figure 7.
Linear correlation between the expression levels of long non-coding RNAs and mRNAs including (A) DLEU2/FBN2, (B) DLEU2/TRIM36, (C) C8orf31/WT1 and (D) MYCNOS/TRIM71.. Cor, correlation coefficient; FBN2, fibrillin 2; TRIM, tripartite motif containing; WT1, WT1 transcription factor; DLEU2, deleted in lymphocytic leukemia 2; C8orf31, chromosome 8 open reading frame 31; MYCNOS, MYCN opposite strand.
Table IV.
Univariate and multivariate Cox regression analysis of overall survival in the TARGET.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variable | P-value | HR | 95% CI | P-value | HR | 95% CI |
| Sex (male vs. female) | 0.0550 | 1.7240 | 0.9880–3.0090 | 0.0710 | 1.7490 | 0.9530–3.2090 |
| Ethnicity (black vs. white) | 0.0380 | 2.0110 | 1.0410–3.8870 | 0.1590 | 1.6450 | 0.8230–3.2890 |
| Age (>2 vs. <2 years) | 0.2070 | 1.8140 | 0.7200–3.5760 | 0.7330 | 1.2050 | 0.4130–3.5130 |
| NMTS Stage (III–V vs. I–II) | <0.0001 | 3.3020 | 1.8190–3.9940 | 0.0030 | 2.8570 | 1.4370–3.6840 |
| Histology (DAWT vs. FHWT) | 0.8380 | 1.0640 | 0.5870–3.9280 | 0.9350 | 0.9680 | 0.4470–3.0990 |
| MYCNOS | 0.0030 | 1.4510 | 1.1330–3.8570 | 0.0420 | 1.3550 | 1.1060–3.8150 |
| DLEU2 | 0.0550 | 1.3500 | 0.9940–3.8360 | 0.8560 | 1.0600 | 0.7220–3.4840 |
| C8orf31 | 0.0500 | 0.7580 | 0.5740–3.0010 | 0.7590 | 0.9530 | 0.6980–3.2990 |
HR, hazard ratio; CI, confidence interval; NMTS, National Wilms Tumor Study; DAWT, diffuse anaplastic histology Wilms tumor; FHWT, favorable histology Wilms tumor; MYCNOS, MYCN opposite strand; DLEU2, deleted in lymphocytic leukemia 2; C8orf31, chromosome 8 open reading frame 31.
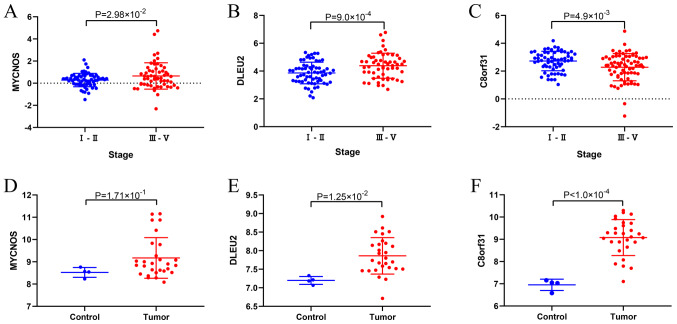
Validation of DElncRNAs
The favorable histology WTs in the TARGET database cohort (Table I) of the present study were obtained from patients with relapse, and therefore, only the differences in the expression of the three lncRNAs in the late and early stages of WT were compared. The results suggested that MYCNOS and DELU2 were highly expressed (P=2.98×10−2, P=9.0×10−4) in the late stages of WT, whereas C8orf31 was highly expressed (P=4.9×10−3) in the early stages of WT (Fig. 8A-C). To validate that the three lncRNAs were also differentially expressed in other datasets, the GSE66405 dataset from GEO was investigated. DLEU2 and C8orf31 were differentially expressed in tumor and normal tissues in the GSE66405 dataset (P=1.25×10−2, P<1.0×10−4; Fig. 8E and F). Additionally, the expression of MYCNOS in the tumor tissues was higher than that in the normal tissues, but this difference was not statistically significant (P=1.71×10−1; Fig. 8D).
Figure 8.
Expression of MYCNOS, DLEU2 and C8orf31. Differential expression of (A) MYCNOS, (B) DLEU2 and (C) C8orf31 in the high and low stages of Wilms tumor in the Therapeutically Applicable Research to Generate Effective Treatments database. Differential expression of (D) MYCNOS, (E) DELU2 and (F) C8orf31 between Wilms tumor and normal kidney tissues in the GSE66405 dataset from the Gene Expression Omnibus database. MYCNOS, MYCN opposite strand; DLEU2, deleted in lymphocytic leukemia 2; C8orf31, chromosome 8 open reading frame 31.
Discussion
WT presents high morbidity and mortality in children; although the overall 5-year survival rate for patients with WT has improved from 20% in the 1960s to >90% in the 2000s (3,4), the current treatments for WT, including surgery, chemotherapy and radiation, still have limitations and complications, including death (31,32). A study involving 6,185 patients with WT reported that the risk of death in WT survivors remained high several years after the original diagnosis (33). Therefore, the identification of novel key molecules as targets for biotherapy would be beneficial for improving the current status of WT treatment.
Recently, lncRNAs have gained increasing attention (14). Different lncRNAs influence several cellular processes, such as development and development, antiviral response and gene imprinting, but the specific function of the majority of lncRNAs is still unknown (34,35). Numerous theories and hypotheses have been proposed to explain how lncRNAs regulate the biological behavior of tumor cells (34,36). The ceRNA hypothesis (19), which states that lncRNAs regulate mRNAs by competing for miRNAs, has been proven (20,37–39). A number of previous studies have investigated ceRNAs in other pediatric malignant tumors (40,41), but there are only a few studies on the role of ceRNAs in WT (23–26). To further understand how lncRNA-related ceRNA networks affect WT, the present study analyzed large-scale sequencing data of WT tissues from the TARGET database.
In the present study, 442 lncRNAs, 214 miRNAs and 4,912 mRNAs were identified as DEGs between normal and WT tissues. Moreover, functional analysis of the DEmRNAs was performed to determine their potential biological mechanisms. The GO enrichment analysis indicated that ‘DNA-dependent DNA replication’, ‘DNA replication’, ‘chromosome segregation’, ‘sister chromatid segregation’, ‘mitotic nuclear division’, ‘nuclear chromosome segregation’, ‘sister chromatid cohesion’, ‘mitotic sister chromatid segregation’, ‘nuclear division’ and ‘kidney development’ were the main biological processes of the DEGs associated with WT. The majority of the identified biological processes were associated with cell mitosis and kidney development, which are closely associated with the occurrence and development of renal tumors (42,43). The KEGG pathway enrichment analysis indicated that the majority of DEmRNAs were enriched in ‘DNA replication’, ‘human T-cell leukemia virus 1 infection’, ‘cell cycle’, ‘lysosome’, ‘cell adhesion molecules’, ‘cellular senescence’, ‘p53 signaling pathway’, ‘Rap1 signaling pathway’, ‘MAPK signaling pathway’ and ‘toxoplasmosis’, with ‘MAPK signaling pathway’ displaying the highest level of significance. A recent study suggested that the mitogen-activated protein kinase/ERK signaling pathway plays a role in WT (44). Among the DEGs, 32 lncRNAs, 14 miRNAs and 158 mRNAs were included in the ceRNA network. Su et al (11) reported that small nucleolar RNA host gene (SNHG)6 decreases the proliferation, migration and invasion of WT cell lines by regulating miR-15a. The SNHG family genes, including SNHG1, SNHG5 and SNHG6, were also present in the ceRNA network of the present study. Their regulatory network with miRNAs may be even more complex (45,46). Furthermore, three DElncRNAs (MYCNOS, DLEU2 and C8orf31), three DEmiRNAs (hsa-miR-135a-3p, hsa-miR-363-3p and hsa-miR-125b-5p) and 17 DEmRNAs (WT1, FAM102A, FOXP4, TRIM36, TRIM71, FBN2, GNS, KLF10, LIMA1, PPP1R3B, PSD3, STAT3, STAT6, TMEM127, UXS1, VLDLR and ZBTB4) were significantly associated with OS. MYCNOS is transcribed in the anti-sense strand across exon1 and intron1 from MYCN (47). The gene is highly expressed in small cell lung cancer, MYCN-amplified rhabdomyosarcoma and neuroblastoma (48,49). Zhao et al (50) reported that MYCNOS cooperated with CCCTC-binding factor to promote the proliferation, invasion and metastasis of neuroblastoma cells in vitro and in vivo. In the present study, MYCNOS was upregulated and associated with poor OS in the TARGET dataset. However, in the GSE66405 dataset, the difference in MYCNOS expression was not statistically significant between the tumor and normal tissues. This difference between results might be due to the small sample size of the present study. DLEU2 has been reported to affect the biological behaviors of multiple types of cancer, such as laryngeal carcinoma and leukemia (51,52). Xie et al (51) reported that DLEU2 can induce the proliferation, migration and invasion of laryngeal carcinoma cells via miR-16-1. The present study also suggested that C8orf31, in the ceRNA network, was highly expressed in WT tissues. However, the high expression of C8orf31 was detected only in early stage tumors and observed in patients with improved survival. This suggested that the upregulation of C8orf31 may be a protective mechanism in WT. As the malignancy of the tumor increased, the expression of C8orf31 decreased. However, based on the present results also a correlation between these factors can be determined, and not a causative effect. Thus, further investigation is required to identify the function of C8orf31 in WT and other tumors.
miRNAs can regulate various target genes, playing an important role in the development of cancer (53,54). In the present study, three miRNAs (hsa-miR-135a-3p, hsa-miR-363-3p and hsa-miR-125b-5p) were identified in the ceRNA network and were positively associated with OS. Previous studies have reported that the three miRNAs act as tumor suppressors in a number of types of cancer (55–61). Fukagawa et al (56) reported that the overexpression of miR-135a-3p enhanced ovarian cancer cell sensitivity to chemotherapy drugs and suppressed proliferation. Li et al (61) reported that miR-363-3p was downregulated in renal cell carcinoma and reduced the inhibition of the oncogenic cyclic adenosine monophosphate responsive element binding protein 1. miR-125b-5p can inhibit γδ T cell activity, promote γδ T cell apoptosis and subsequently suppress the immune response to infections and tumorigenesis (62). Additionally, 17 DEmRNAs in the ceRNA network were identified and were significantly related to the OS rate, including WT1, which is closely related to the development of WT (63). Certain well-known genes, including FOXP4 and STAT3/6, play a role in tumorigenesis but not in WT, and certain less reported genes, including FAM102A and FBN2, require further investigation (64,65). In the present study, MYCNOS and TRIM71 displayed the highest linear correlation. A number of previous studies have demonstrated that high levels of TRIM71, also known as LIN41, are associated with hepatocellular carcinoma (66) and myxoid liposarcoma (67), while another study reported that TRIM71 inhibited tumorigenesis via the regulation of the Lin28B-let-7-high mobility group AT-hook 2 signaling pathway in colorectal carcinoma cells and non-small cell lung cancer cells (68). Therefore, further investigation is required to identify the roles of various genes and molecular mechanisms underlying WT. Although the results of the present study were validated using another dataset, a potential limitation of the present study was the small sample size, therefore further investigation using larger sample sizes is required.
To conclude, the present study investigated the expression of lncRNAs, miRNAs and mRNAs specific to WT and their potential functions. A ceRNA network was constructed to provide a novel insight into the role of lncRNAs in the development of WT. The three lncRNAs identified in the ceRNA network may aid in the prognosis of WT and serve as potential targets for clinical therapy.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the TARGET (https://ocg.cancer.gov/programs/target/data-matrix) and GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE66405) repository.
Authors' contributions
DS was involved in the conception of the study. ZW designed and drafted the manuscript. ZW, LQ, HC and DS collected and analyzed the data. HC was involved in interpretation of data. ZW, HC, LQ and DS revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bernstein L, Linet M, Smith MA, editors. National Cancer Institute; Bethesda, MD: 1999. Cancer Incidence and Survival Among Children and Adolescents: United States SEER Program, 1975-1995; p. p79. [Google Scholar]
- 2.Grovas A, Fremgen A, Rauck A, Ruymann FB, Hutchinson CL, Winchester DP, Menck HR. The National Cancer Data Base report on patterns of childhood cancers in the United States. Cancer. 1997;80:2321–2332. doi: 10.1002/(SICI)1097-0142(19971215)80:12<2321::AID-CNCR14>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 3.Koshinaga T, Takimoto T, Oue T, Okita H, Tanaka Y, Nozaki M, Tsuchiya K, Inoue E, Haruta M, Kaneko Y, et al. Outcome of renal tumors registered in Japan Wilms Tumor Study-2 (JWiTS-2): A report from the Japan Children's Cancer Group (JCCG) Pediatr Blood Cancer. 2018;65:e27056. doi: 10.1002/pbc.27056. [DOI] [PubMed] [Google Scholar]
- 4.Metzger ML, Dome JS. Current therapy for Wilms' tumor. Oncologist. 2005;10:815–826. doi: 10.1634/theoncologist.10-10-815. [DOI] [PubMed] [Google Scholar]
- 5.Smith GR, Thomas PR, Ritchey M, Norkool P. Long-term renal function in patients with irradiated bilateral Wilms tumor. National Wilms' Tumor Study Group. Am J Clin Oncol. 1998;21:58–63. doi: 10.1097/00000421-199802000-00013. [DOI] [PubMed] [Google Scholar]
- 6.Green DM, Grigoriev YA, Nan B, Takashima JR, Norkool PA, D'Angio GJ, Breslow NE. Congestive heart failure after treatment for Wilms' tumor: A report from the National Wilms' Tumor Study group. J Clin Oncol. 2001;19:1926–1934. doi: 10.1200/JCO.2001.19.7.1926. [DOI] [PubMed] [Google Scholar]
- 7.Green DM, Norkool P, Breslow NE, Finklestein JZ, D'Angio GJ. Severe hepatic toxicity after treatment with vincristine and dactinomycin using single-dose or divided-dose schedules: A report from the National Wilms' Tumor Study. J Clin Oncol. 1990;8:1525–1530. doi: 10.1200/JCO.1990.8.9.1525. [DOI] [PubMed] [Google Scholar]
- 8.Green DM, Peabody EM, Nan B, Peterson S, Kalapurakal JA, Breslow NE. Pregnancy outcome after treatment for Wilms tumor: A report from the National Wilms Tumor Study Group. J Clin Oncol. 2002;20:2506–2513. doi: 10.1200/JCO.2002.07.159. [DOI] [PubMed] [Google Scholar]
- 9.van Dijk IW, Oldenburger F, Cardous-Ubbink MC, Geenen MM, Heinen RC, de Kraker J, van Leeuwen FE, van der Pal HJ, Caron HN, Koning CC, et al. Evaluation of late adverse events in long-term Wilms' tumor survivors. Int J Radiat Oncol Biol Phys. 2010;78:370–378. doi: 10.1016/j.ijrobp.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 10.Howlader N, Noone AM, Krapcho M, Miller D, Bishop K, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, et al., editors. NIH publication National Cancer Institute; Bethesda, MD: 2017. SEER Cancer Statistics Review, 1975–2014; p. p996. [Google Scholar]
- 11.International Human Genome Sequencing Consortium, corp-author. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 12.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang K, Lu C, Huang X, Cui J, Li J, Gao Y, Liang W, Liu Y, Sun Y, Liu H, et al. Long noncoding RNA AOC4P regulates tumor cell proliferation and invasion by epithelial-mesenchymal transition in gastric cancer. Therap Adv Gastroenterol. 2019;12:1756284819827697. doi: 10.1177/1756284819827697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tran DDH, Kessler C, Niehus SE, Mahnkopf M, Koch A, Tamura T. Myc target gene, long intergenic noncoding RNA, Linc00176 in hepatocellular carcinoma regulates cell cycle and cell survival by titrating tumor suppressor microRNAs. Oncogene. 2018;37:75–85. doi: 10.1038/onc.2017.312. [DOI] [PubMed] [Google Scholar]
- 17.Wang M, Guo C, Wang L, Luo G, Huang C, Li Y, Liu D, Zeng F, Jiang G, Xiao X. Long noncoding RNA GAS5 promotes bladder cancer cells apoptosis through inhibiting EZH2 transcription. Cell Death Dis. 2018;9:238. doi: 10.1038/s41419-018-0264-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu H, Zheng JF, Hou CZ, Li Y, Liu PS. Up-regulation of long intergenic noncoding RNA 01296 in ovarian cancer impacts invasion, apoptosis and cell cycle distribution via regulating EMT. Cell Signal. 2019;62:109341. doi: 10.1016/j.cellsig.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong X, Jin Z, Chen Y, Xu H, Ma C, Hong X, Li Y, Zhao G. Knockdown of long non-coding RNA ANRIL inhibits proliferation, migration, and invasion but promotes apoptosis of human glioma cells by upregulation of miR-34a. J Cell Biochem. 2018;119:2708–2718. doi: 10.1002/jcb.26437. [DOI] [PubMed] [Google Scholar]
- 21.Qu Z, Li S. Long noncoding RNA LINC01278 favors the progression of osteosarcoma via modulating miR-133a-3p/PTHR1 signaling. J Cell Physiol. 2020 Jan 29; doi: 10.1002/jcp.29582. (Epub ahead of print). doi: 10.1002/jcp.29582. [DOI] [PubMed] [Google Scholar]
- 22.Xu W, Yu S, Xiong J, Long J, Zheng Y, Sang X. CeRNA regulatory network-based analysis to study the roles of noncoding RNAs in the pathogenesis of intrahepatic cholangiocellular carcinoma. Aging (Albany NY) 2020;12:1047–1086. doi: 10.18632/aging.102634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su L, Wu A, Zhang W, Kong X. Silencing long non-coding RNA SNHG6 restrains proliferation, migration and invasion of Wilms' tumour cell lines by regulating miR-15a. Artif Cells Nanomed Biotechnol. 2019;47:2670–2677. doi: 10.1080/21691401.2019.1633338. [DOI] [PubMed] [Google Scholar]
- 24.Zhu KR, Sun QF, Zhang YQ. Long non-coding RNA LINP1 induces tumorigenesis of Wilms' tumor by affecting Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:5691–5698. doi: 10.26355/eurrev_201907_18306. [DOI] [PubMed] [Google Scholar]
- 25.Liu Z, He F, OuYang S, Li Y, Ma F, Chang H, Cao D, Wu J. miR-140-5p could suppress tumor proliferation and progression by targeting TGFBRI/SMAD2/3 and IGF-1R/AKT signaling pathways in Wilms' tumor. BMC Cancer. 2019;19:405. doi: 10.1186/s12885-019-5609-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu S, Fu W, Zhang L, Fu K, Hu J, Jia W, Liu G. LINC00473 antagonizes the tumour suppressor miR-195 to mediate the pathogenesis of Wilms tumour via IKKalpha. Cell Prolif. 2018 Feb 51; doi: 10.1111/cpr.12416. (Epub ahead of print). doi: 10.1111/cpr.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludwig N, Werner TV, Backes C, Trampert P, Gessler M, Keller A, Lenhof HP, Graf N, Meese E. Combining miRNA and mRNA expression profiles in Wilms tumor subtypes. Int J Mol Sci. 2016;17:475. doi: 10.3390/ijms17040475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ritchey ML, Shamberger RC, Haase G, Horwitz J, Bergemann T, Breslow NE. Surgical complications after primary nephrectomy for Wilms' tumor: Report from the National Wilms' Tumor Study Group. J Am Coll Surg. 2001;192:63–68, quiz 146. doi: 10.1016/S1072-7515(00)00749-3. [DOI] [PubMed] [Google Scholar]
- 32.Termuhlen AM, Tersak JM, Liu Q, Yasui Y, Stovall M, Weathers R, Deutsch M, Sklar CA, Oeffinger KC, Armstrong G, et al. Twenty-five year follow-up of childhood Wilms tumor: A report from the Childhood Cancer Survivor Study. Pediatr Blood Cancer. 2011;57:1210–1216. doi: 10.1002/pbc.23090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cotton CA, Peterson S, Norkool PA, Takashima J, Grigoriev Y, Green DM, Breslow NE. Early and late mortality after diagnosis of wilms tumor. J Clin Oncol. 2009;27:1304–1309. doi: 10.1200/JCO.2008.18.6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:206. doi: 10.1186/s13059-017-1348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJ, Gough J, Denisenko E, Schmeier S, Poulsen TM, Severin J, et al. An atlas of human long non-coding RNAs with accurate 5 ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 37.Wang L, Wang C, Wu T, Sun F. Long non-coding RNA TP73-AS1 promotes TFAP2B-mediated proliferation, metastasis and invasion in retinoblastoma via decoying of miRNA-874-3p. J Cell Commun Signal. 2020 Feb 18; doi: 10.1007/s12079-020-00550-x. (Epub ahead of print). doi: 10.1007/s12079-020-00550-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Wang J, Cao Y, Lu X, Wang X, Kong X, Bo C, Li S, Bai M, Jiao Y, Gao H, et al. Identification of the regulatory role of lncRNA SNHG16 in myasthenia gravis by constructing a competing endogenous RNA Network. Mol Ther Nucleic Acids. 2020;19:1123–1133. doi: 10.1016/j.omtn.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu J, Zhang J, Shan F, Wen J, Wang Y. SSTR5 AS1 functions as a ceRNA to regulate CA2 by sponging miR 15b 5p for the development and prognosis of HBV related hepatocellular carcinoma. Mol Med Rep. 2019;20:5021–5031. doi: 10.3892/mmr.2019.10736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chi R, Chen X, Liu M, Zhang H, Li F, Fan X, Wang W, Lu H. Role of SNHG7-miR-653-5p-STAT2 feedback loop in regulating neuroblastoma progression. J Cell Physiol. 2019;234:13403–13412. doi: 10.1002/jcp.28017. [DOI] [PubMed] [Google Scholar]
- 41.Fei D, Zhang X, Liu J, Tan L, Xing J, Zhao D, Zhang Y. Long noncoding RNA FER1L4 suppresses tumorigenesis by regulating the expression of PTEN targeting miR-18a-5p in osteosarcoma. Cell Physiol Biochem. 2018;51:1364–1375. doi: 10.1159/000495554. [DOI] [PubMed] [Google Scholar]
- 42.Arai E, Gotoh M, Tian Y, Sakamoto H, Ono M, Matsuda A, Takahashi Y, Miyata S, Totsuka H, Chiku S, et al. Alterations of the spindle checkpoint pathway in clinicopathologically aggressive CpG island methylator phenotype clear cell renal cell carcinomas. Int J Cancer. 2015;137:2589–2606. doi: 10.1002/ijc.29630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hohenstein P, Pritchard-Jones K, Charlton J. The yin and yang of kidney development and Wilms' tumors. Genes Dev. 2015;29:467–482. doi: 10.1101/gad.256396.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurtzeborn K, Kwon HN, Kuure S. MAPK/ERK signaling in regulation of renal differentiation. Int J Mol Sci. 2019;20:E1779. doi: 10.3390/ijms20071779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie M, Zhang Z, Cui Y. Long Noncoding RNA SNHG1 contributes to the promotion of prostate cancer cells through regulating miR-377-3p/AKT2 axis. Cancer Biother Radiopharm. 2020;35:109–119. doi: 10.1089/cbr.2019.3177. [DOI] [PubMed] [Google Scholar]
- 46.Meng X, Deng Y, Lv Z, Liu C, Guo Z, Li Y, Liu H, Xie B, Jin Z, Lin F, et al. LncRNA SNHG5 promotes proliferation of glioma by regulating miR-205-5p/ZEB2 axis. OncoTargets Ther. 2019;12:11487–11496. doi: 10.2147/OTT.S228439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krystal GW, Armstrong BC, Battey JF. N-myc mRNA forms an RNA-RNA duplex with endogenous antisense transcripts. Mol Cell Biol. 1990;10:4180–4191. doi: 10.1128/MCB.10.8.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Armstrong BC, Krystal GW. Isolation and characterization of complementary DNA for N-cym, a gene encoded by the DNA strand opposite to N-myc. Cell Growth Differ. 1992;3:385–390. [PubMed] [Google Scholar]
- 49.O'Brien EM, Selfe JL, Martins AS, Walters ZS, Shipley JM. The long non-coding RNA MYCNOS-01 regulates MYCN protein levels and affects growth of MYCN-amplified rhabdomyosarcoma and neuroblastoma cells. BMC Cancer. 2018;18:217. doi: 10.1186/s12885-018-4129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao X, Li D, Pu J, Mei H, Yang D, Xiang X, Qu H, Huang K, Zheng L, Tong Q. CTCF cooperates with noncoding RNA MYCNOS to promote neuroblastoma progression through facilitating MYCN expression. Oncogene. 2016;35:3565–3576. doi: 10.1038/onc.2015.422. [DOI] [PubMed] [Google Scholar]
- 51.Xie ZZ, Xiao ZC, Song YX, Li W, Tan GL. Long non-coding RNA Dleu2 affects proliferation, migration and invasion ability of laryngeal carcinoma cells through triggering miR-16-1 pathway. Eur Rev Med Pharmacol Sci. 2018;22:1963–1970. doi: 10.26355/eurrev_201804_14723. [DOI] [PubMed] [Google Scholar]
- 52.Lerner M, Harada M, Lovén J, Castro J, Davis Z, Oscier D, Henriksson M, Sangfelt O, Grandér D, Corcoran MM. DLEU2, frequently deleted in malignancy, functions as a critical host gene of the cell cycle inhibitory microRNAs miR-15a and miR-16-1. Exp Cell Res. 2009;315:2941–2952. doi: 10.1016/j.yexcr.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 53.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: Progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 55.Duan S, Dong X, Hai J, Jiang J, Wang W, Yang J, Zhang W, Chen C. MicroRNA-135a-3p is downregulated and serves as a tumour suppressor in ovarian cancer by targeting CCR2. Biomed Pharmacother. 2018;107:712–720. doi: 10.1016/j.biopha.2018.08.044. [DOI] [PubMed] [Google Scholar]
- 56.Fukagawa S, Miyata K, Yotsumoto F, Kiyoshima C, Nam SO, Anan H, Katsuda T, Miyahara D, Murata M, Yagi H, et al. MicroRNA-135a-3p as a promising biomarker and nucleic acid therapeutic agent for ovarian cancer. Cancer Sci. 2017;108:886–896. doi: 10.1111/cas.13210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang SH, Zhang WJ, Wu XC, Weng MZ, Zhang MD, Cai Q, Zhou D, Wang JD, Quan ZW. The lncRNA MALAT1 functions as a competing endogenous RNA to regulate MCL-1 expression by sponging miR-363-3p in gallbladder cancer. J Cell Mol Med. 2016;20:2299–2308. doi: 10.1111/jcmm.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dong J, Geng J, Tan W. MiR-363-3p suppresses tumor growth and metastasis of colorectal cancer via targeting SphK2. Biomed Pharmacother. 2018;105:922–931. doi: 10.1016/j.biopha.2018.06.052. [DOI] [PubMed] [Google Scholar]
- 59.Wang Y, Chen T, Huang H, Jiang Y, Yang L, Lin Z, He H, Liu T, Wu B, Chen J, et al. miR-363-3p inhibits tumor growth by targeting PCNA in lung adenocarcinoma. Oncotarget. 2017;8:20133–20144. doi: 10.18632/oncotarget.15448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu J, Li Q, Li R, Ren P, Dong S. MicroRNA-363-3p inhibits papillary thyroid carcinoma progression by targeting PIK3CA. Am J Cancer Res. 2017;7:148–158. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Li Y, Wang Y, Fan H, Zhang Z, Li N. miR-125b-5p inhibits breast cancer cell proliferation, migration and invasion by targeting KIAA1522. Biochem Biophys Res Commun. 2018;504:277–282. doi: 10.1016/j.bbrc.2018.08.172. [DOI] [PubMed] [Google Scholar]
- 62.Zhu Y, Zhang S, Li Z, Wang H, Li Z, Hu Y, Chen H, Zhang X, Cui L, Zhang J, et al. miR-125b-5p and miR-99a-5p downregulate human γδ T-cell activation and cytotoxicity. Cell Mol Immunol. 2019;16:112–125. doi: 10.1038/cmi.2017.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee SB, Haber DA. Wilms tumor and the WT1 gene. Exp Cell Res. 2001;264:74–99. doi: 10.1006/excr.2000.5131. [DOI] [PubMed] [Google Scholar]
- 64.Kim JH, Hwang J, Jung JH, Lee HJ, Lee DY, Kim SH. Molecular networks of FOXP family: Dual biologic functions, interplay with other molecules and clinical implications in cancer progression. Mol Cancer. 2019;18:180. doi: 10.1186/s12943-019-1110-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Verhoeven Y, Tilborghs S, Jacobs J, De Waele J, Quatannens D, Deben C, Prenen H, Pauwels P, Trinh XB, Wouters A, et al. The potential and controversy of targeting STAT family members in cancer. Semin Cancer Biol. 2020;60:41–56. doi: 10.1016/j.semcancer.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 66.Chen YL, Yuan RH, Yang WC, Hsu HC, Jeng YM. The stem cell E3-ligase Lin-41 promotes liver cancer progression through inhibition of microRNA-mediated gene silencing. J Pathol. 2013;229:486–496. doi: 10.1002/path.4130. [DOI] [PubMed] [Google Scholar]
- 67.De Cecco L, Negri T, Brich S, Mauro V, Bozzi F, Dagrada G, Disciglio V, Sanfilippo R, Gronchi A, D'Incalci M, et al. Identification of a gene expression driven progression pathway in myxoid liposarcoma. Oncotarget. 2014;5:5965–5977. doi: 10.18632/oncotarget.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yin J, Kim TH, Park N, Shin D, Choi HI, Cho S, Park JB, Kim JH. TRIM71 suppresses tumorigenesis via modulation of Lin28B-let-7-HMGA2 signaling. Oncotarget. 2016;7:79854–79868. doi: 10.18632/oncotarget.13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the TARGET (https://ocg.cancer.gov/programs/target/data-matrix) and GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE66405) repository.