Abstract
O-Linked N-acetyl glucosamine (O-GlcNAc) is a protein modification found on thousands of nuclear, cytosolic, and mitochondrial proteins. Many O-GlcNAc sites occur in close proximity to protein sites that are likewise modified by phosphorylation. While several studies have uncovered crosstalk between these two signaling modifications on individual proteins and pathways, an understanding of the role of O-GlcNAc in regulating kinases, the enzymes that install the phosphate modification, is still emerging. Here we review recent methods to profile the O-GlcNAc modification on a global scale that have revealed over 100 kinases as modified by O-GlcNAc, and highlight existing studies about regulation of these kinases by O-GlcNAc. Continuing efforts to profile the O-GlcNAc proteome and understand the role of O-GlcNAc on kinases will reveal new mechanisms of regulation and potential avenues for manipulation of the signaling mechanisms at the intersection of O-GlcNAc and phosphorylation.
Keywords: O-GlcNAc, glycosylation, kinase, phosphatase, phosphorylation, glycoproteomics, crosstalk, post-translational modification, PTM crosstalk, signaling
Introduction
O-Linked N-acetyl glucosamine (O-GlcNAc) is one of the major chemical codes used for cellular signaling. O-GlcNAc was first discovered in mammalian cells in 19841 and has now been found across species on serine or threonine residues of thousands of proteins in the nuclear, cytosolic, and mitochondrial compartments (Figure 1A).2, 3 Due to the occurrence of O-GlcNAc on proteins in these intracellular compartments and the fact that O-GlcNAc is a biosynthetic product culminating from glycolysis, amino acid synthesis, nucleotide levels, and fatty acid levels, the modification is commonly thought to act as a nutrient sensor for the proteome.3 The O-GlcNAc modification is found across species. The O-GlcNAc modification modifies thousands of proteins in animals and plants,4 and has now been identified on proteins in fungi5 and the tailoring enzymes has been found in bacteria.6 Notably, the recent discovery of intracellular O-fucose in the plant proteome7 and O-mannose in the yeast proteome8 points to the potentially central role for a sugar-based nutrient sensing mechanism across organisms. The function of O-GlcNAc in nutrient sensing and the impact of its dysregulation on specific diseases have been previously and extensively reviewed,9–17 which highlights a rapidly growing focus on deciphering the O-GlcNAc code.
Figure 1.

The essential O-GlcNAc modification of proteins. A. Structure of O-GlcNAc (highlighted in red) and phosphate appended to a serine or threonine amino acid. B. O-GlcNAc is installed by OGT and removed by OGA to over 3,000 known nuclear and cytoplasmic proteins.
The intersection between O-GlcNAc and phosphorylation signaling was first posited with the discovery of O-GlcNAc on serine and threonine residues that also serve as modification sites for phosphorylation.18 This discovery sparked multiple inquiries aimed at investigating the crosstalk between these two modifications. The interface of O-GlcNAc and phosphorylation has since proven to be an important pillar of cellular signaling arising from these modifications, particularly as many regions of O-GlcNAcylation are also substrates for phosphorylation. O-GlcNAc, like phosphorylation, is a dynamic modification that is rapidly cycled on protein substrates.15 However, in contrast to phosphorylation for which over 500 kinases and phosphatases execute the enzymatic introduction and removal of the phosphate group,19 O-GlcNAc is tailored on thousands of substrates by only two enzymes: O-GlcNAc transferase (OGT) installs and O-GlcNAcase (OGA) removes O-GlcNAc from proteins (Figure 1B). Evidence for the functional significance of O-GlcNAc cycling was obtained when early work demonstrated that both OGT and OGA are essential for organismal development. OGT is required for normal human neurological development20 and deletion of OGT leads to embryonic lethality in mice,21 while deletion of OGA leads to neonatal lethality with developmental delay in mouse embryos.22 Furthermore, conditional deletion of OGT in numerous cell types leads to senescence and apoptosis.23 Numerous functional outcomes of protein O-GlcNAcylation have been described, including alteration of protein–protein interactions, subcellular localization, enzymatic activity, and protein stability.15, 24 The dysregulation and abnormal levels of O-GlcNAc have been linked to various diseases, including diabetes,25 cancer,26, 27 immune disorders28 and neurodegeneration,29, 30 which have inspired efforts to pursue the O-GlcNAc modification as a potential therapeutic target.31, 32
Signaling between O-GlcNAc and phosphorylation pathways may be broadly categorized into two major models: that of crosstalk on a protein substrate, or through post-translational regulation of the PTM-installing enzymes themselves (Figure 2). In the first model, a protein substrate may act as a scaffold for O-GlcNAc or phosphorylation via competitive modification or cooperative modification (Figure 2A). In instances of competitive modification, a protein substrate may be alternatively glycosylated or phosphorylated, which inhibits the subsequent modification of the protein at the same or proximal amino acid residues. The competitive modification of a protein by O-GlcNAc or phosphorylation has been described for the tumor suppressor p53,33 the oncoprotein c-Myc,34 and the Alzheimer’s associated-protein tau,35 among others. For example, the structural effects of modification on a region of tau revealed that the addition of phosphate drove helical formation, while the addition of O-GlcNAc opposed helical formation by NMR solution structure.36 Conversely, modification of the murine estrogen receptor with O-GlcNAc increased the helical turn propensity while phosphorylation decreased helicity.37 In instances of cooperative modification, the O-GlcNAcylation or phosphorylation of a protein substrate promotes subsequent modification of the protein substrate. For example, mutation of the three O-GlcNAc sites of cyclin-dependent kinase inhibitor p27(Kip1) to alanine significantly decreased phosphorylation at S10; conversely, a S10A mutation decreased O-GlcNAcylation of p27(Kip1), while the S10E mutation acted as a phosphomimetic that increased O-GlcNAcylation of p27(Kip1).38 Recently, the 14-3-3 proteins were identified as receptors for the O-GlcNAc modification, pointing to the potential integration of O-GlcNAc and phosphorylation signaling pathways through a common protein “reader”.39 Thus, tuning a protein surface with O-GlcNAc, phosphorylation, or a combination thereof results in distinct conformational changes that drive downstream functional effects; these have been previously reviewed.10, 40, 41
Figure 2.
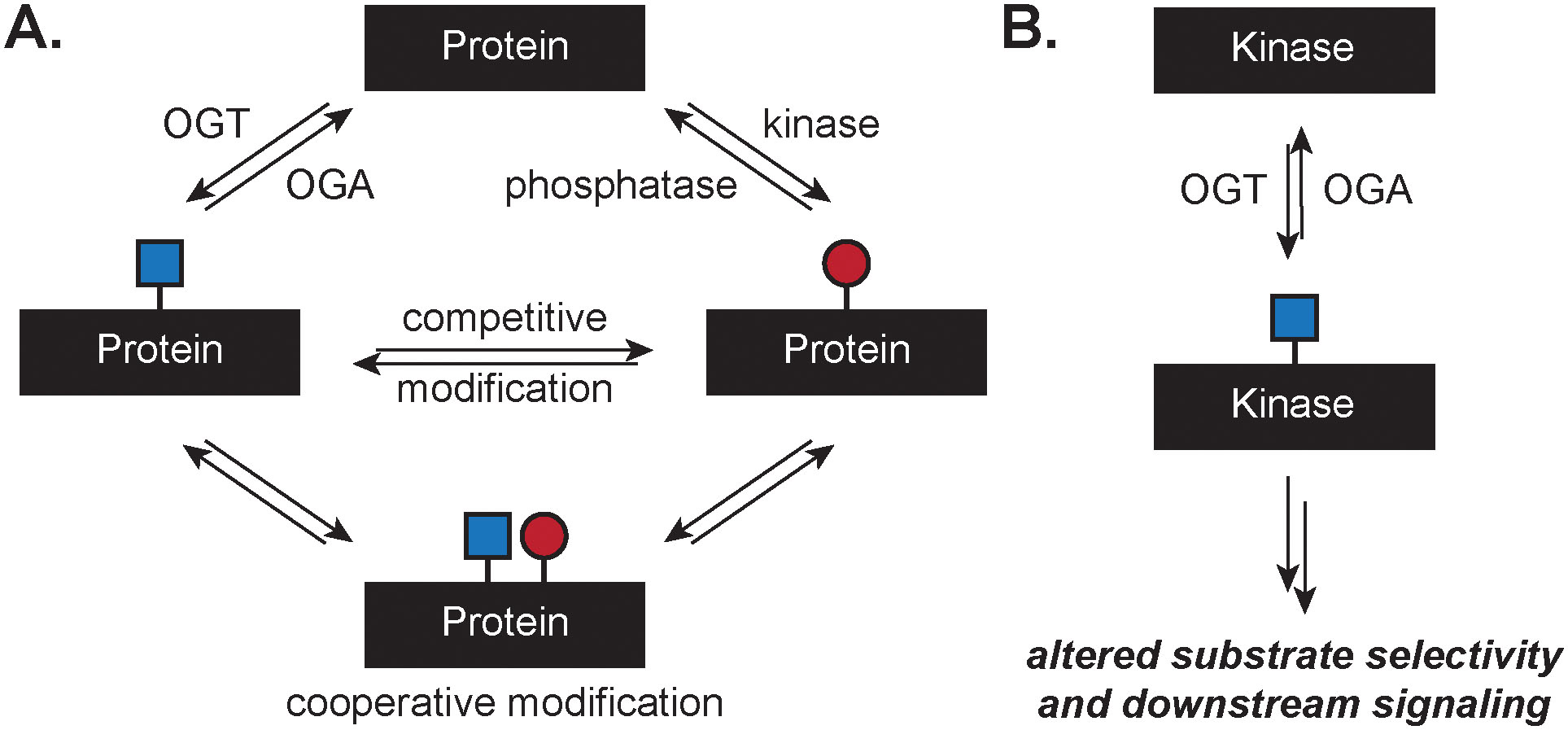
Modes of crosstalk between O-GlcNAc and phosphorylation. A. Post-translational modification of a protein with O-GlcNAc or phosphorylation may be competitive, where one modification precludes the other, or cooperative, where multiple modifications propagate specific regulatory outcomes. B. Modification of a kinase with O-GlcNAc can alter downstream substrate selection and signaling through phosphorylation.
Here, we aim to highlight the intersection of O-GlcNAc and phosphorylation from studies on the systems level and focus on an additional model arising from the system-wide profiling data, where the direct modification of kinases by O-GlcNAc results in regulation of substrate scope, kinase activity, and ultimately drives changes to downstream signaling (Figure 2B).
Regulation of O-GlcNAc by O-GlcNAc transferase and O-GlcNAcase
Like the O-GlcNAc modification itself, the enzymes OGT and OGA that install O-GlcNAc perform multifaceted roles that are still under investigation. OGT is a glycosyltransferase that is expressed in all mammalian tissues and possesses a tetratricopeptide repeat (TPR) domain and a catalytic domain (Figure 3). OGT is expressed in three isoforms that vary by the length of the TPR domain, termed the nucleocytoplasmic or full-length OGT (ncOGT, 13.5 TPRs), the mitochondrial isoform (mOGT, 9 TPRs), and the short isoform (sOGT, 2.5 TPRs) (Figure 3A).42 The TPR domain mediates protein–protein interactions (PPIs) of OGT with substrate proteins, which is only beginning to be understood.24, 43–45 Structurally, the TPR domain forms a series of stacked alpha-helical domains that form a coiled tube-like structure that funnels polypeptide sequences to the catalytic domain primarily through associations with asparagine and aspartate residues lining the domain.43, 46 The catalytic domain transfers the sugar from a UDP-GlcNAc donor to serine or threonine residues that are positioned by the TPR domain (Figure 3B). Mutation of H508 or K852 reduce binding of UDP-GlcNAc and thus impair catalytic activity (highlighted in red, Figure 3B). The catalytic domain catalyzes additional chemistries, including transfer of UDP-GlcNAc to cysteine residues,47 transfer of glucose,48 proteolysis,49 and deamidation.50 In cells, OGT forms dimers and trimers through associations in the TPR domain that also alter UDP-GlcNAc binding constants.51, 52 OGT possesses three of its own O-GlcNAc sites and is modified by other PTMs, including phosphorylation, ubiquitinylation, and sumoylation.53 OGT appears to require accessory proteins to modify protein substrates efficiently, as reduction or removal of the TPR domain reduces activity with full length proteins, but retains catalytic activity with synthetic peptides in vitro.42, 46 Further exploration of the substrate selection mechanisms for OGT will improve the understanding of the regulatory role of O-GlcNAc and its dynamic cycling.
Figure 3.
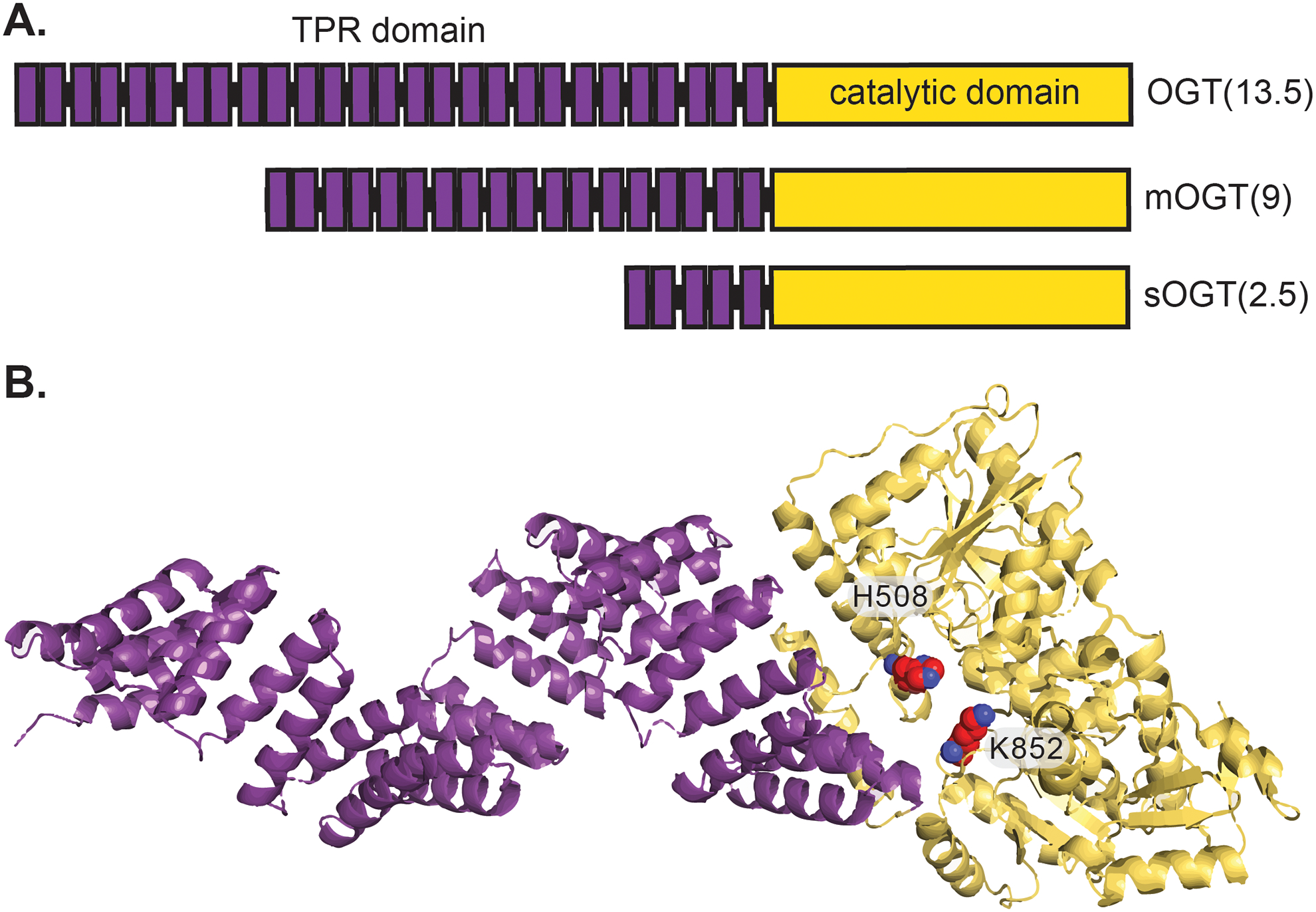
Structure of OGT. A. Linear representation of full length OGT(13.5), mOGT(9), and sOGT(2.5). B. Model of the TPR domain (purple) and catalytic domain of OGT (yellow).43 Point mutations at H508 and K852 reduce catalytic activity (highlighted in red).
O-GlcNAc is catalytically removed from proteins by OGA. Similar to OGT, the mechanisms of how OGA selects from numerous O-GlcNAc substrates and the functions of the separate domains of OGA are still emerging. OGA is composed of three domains: a catalytic domain and a histone acetyltransferase (HAT)-like domain connected by a stalk domain (Figure 4A).54 The stalk domain is interspersed with an unstructured region that forms a binding interaction with OGT.55 OGA is encoded by a single gene that is expressed as two main isoforms in vertebrates, the full length isoform, OGA(I), and a short isoform lacking the C-terminal HAT-like domain, OGA(II).56 Full-length OGA(I) is a nucleocytoplasmic enzyme, while OGA(II) is found predominantly in the nucleus and in lipid droplets.57 The crystal structure of human OGA had eluded definition until very recently.58–60 These structures of OGA(II) revealed a remarkable homodimer, wherein the stalk domain of one monomer covers the catalytic domain of the other monomer to create a substrate-binding cleft.59 A series of contacts between the stalk domains stabilize the homodimer. OGA is regulated at least in part by an O-GlcNAc-related feedback mechanism that triggers gene expression; inhibition of OGA by Thiamet G treatment causes a compensatory increase in OGA expression and decrease in OGT expression.61 Large-scale profiling studies have revealed several sites for ubiquitination and phosphorylation of OGA, including an O-GlcNAc site at S405, indicating possible regulation by OGT.62 The biochemical characterization of these modification sites may illuminate additional mechanisms of OGA regulation.
Figure 4.

Structure of OGA isoforms. A. Linear representation of OGA isoforms I and II. Isoform I is full length OGA. Isoform II lacks the HAT-like domain. B. Crystal structure of the human OGA homodimer analogous to OGA(II) from the side view (PDB: 5UN9).59 The catalytic domain is grey and the stalk domain blue.
Methods to detect and map O-GlcNAc on the systems-level
The O-GlcNAc modification is relatively difficult to detect and quantify in the proteome.2 O-GlcNAc was discovered in 1984 when sugar radiolabeling was measured in the nucleocytoplasmic space,1 several decades after other PTMs like phosphorylation were discovered on the same proteins. The discovery of O-GlcNAc may have been delayed due to difficulty in detection since generally changes in O-GlcNAc levels do not affect glycoprotein migration during gel electrophoresis, and O-GlcNAc is enzymatically labile and rapidly removed from proteins when the cell is damaged or lysed. Furthermore, O-GlcNAc is also chemically labile to common mapping techniques to analyze PTMs, such as mass spectrometry (MS).63 Analysis of O-GlcNAc by MS pushes detection limits due to occurrence of O-GlcNAc at substoichiometric levels on the protein, ion suppression of the glycopeptide in the presence of unmodified peptides, and ready fragmentation of the glycan from the peptide during ionization processes in the mass spectrometer.64, 65 Recently, advances in chemical glycoproteomics have drastically accelerated the mapping of the modification site in the global proteome; these advances have recently been reviewed.66 Here we highlight specifically advances in the large-scale profiling of O-GlcNAc and summarize the O-GlcNAcome characterized from large-scale complex glycoproteomic studies to date.
Although O-GlcNAc is found widely throughout the nucleocytoplasmic proteome, its substoichiometric modification site occupancy necessitates the combination of an efficient enrichment method coupled to a sensitive analytical detection method. The enrichment of O-GlcNAc has been achieved by several means. Lectin weak affinity column chromatography using wheat germ agglutinin (WGA) enables the enrichment of O-GlcNAc and other sugars after multiple rounds of enrichment. Alternatively, the introduction of bioorthogonal handles via metabolic labeling67, 68 or chemoenzymatic labeling69 results in the selective installation of an azido-sugar as a reporter for O-GlcNAc on proteins, enabling the further functionalization with a variety of reporting strategies (e.g., fluorescence microscopy, anti-biotin Western blot). Metabolic labeling involves the addition of a sugar carrying a bioorthogonal handle, such as an azido-sugar, to living systems that metabolically incorporate the azido-sugar to protein substrates. Several sugar reporter molecules for O-GlcNAc have been developed, including Ac4GalNAz or Ac4GlcNAz, or O-GlcNAc-specific reporters 6AlkGlcNAc,70 6AzGlcNAc,71 1,3-Ac2GalNAz, and 1,3-Pr2GalNAz.72 The latter two were validated to reduce background S-glycosylation from other metabolic reporters.73 Chemoenzymatic labeling uses a mutant GalT1 enzyme that accepts azido- or keto-sugars for enzymatic labeling of the O-GlcNAc residue itself.74 The azide introduces a handle that is selectively tagged with reporter molecules using copper-catalyzed azide–alkyne cycloaddition (CuAAC) chemistry. Reaction of the azide groups with cleavable biotin tags and isolation of the O-GlcNAc peptide enables maps of O-GlcNAc modification sites throughout the proteome. Both metabolic labeling and chemoenzymatic labeling may produce off-target labeling products due to the addition of reactive azido-sugar intermediates or promiscuity of the labeling for additional glycan types.73, 75 Thus, the assignment of O-GlcNAcylated proteins is best performed at glycosite-level resolution.
Analysis of the O-GlcNAc modification is commonly achieved by Western blot or MS-based proteomics. Visualization of O-GlcNAcylated proteins by Western blot is commonly performed using the O-GlcNAc CTD110.6, RL2, or 18B10.C7 antibodies. If the glycoprotein is labeled by an azido-sugar, a mass shift assay using a 5-kilodalton PEG mass tag carrying an alkynyl functional group may be performed to determine the stoichiometry of the O-GlcNAc modification on individual proteins.76 The intensity of the shifted bands relative to the unshifted band allows for determination of O-GlcNAc stoichiometry. To characterize the protein glycosite(s), mass spectrometry-based proteomics has emerged as the primary mechanism for site-specific mapping of the O-GlcNAc modification site on individual proteins to the complex proteome. However, due to the chemical lability of the O-linked glycosidic bond from the peptide backbone by collision induced dissociation (CID) or higher-energy CID (HCD) resulting in altered fragmentation mechanisms for glycopeptides, the assignment of the glycopeptide species is challenging, and in cases of successful identification of the glycopeptide the glycosite may only be localized to the serine and threonine residues in the peptide sequence. Solutions to this challenge included determination of the modification site using a sequence of induced beta-elimination of O-GlcNAc from the peptide backbone, followed by controlled Michael addition of dithiothreitol as a reporter for the glycosite, yielding early insight to the O-GlcNAc proteome.77 The further development of ETD and electron-transfer higher energy collision induced dissociation (EthCD) methods on high resolution mass spectrometers enabled the detection of O-GlcNAcylated peptides via a fragmentation method that leaves the glycosidic bond intact. Chemical glycoproteomics methods, such as Isotope Targeted Glycoproteomics (IsoTaG), combine metabolic labeling with enrichment to map exactly when and where O-GlcNAc is modifying the protein network (Figure 5).78 The development of efficient enrichment methods for O-GlcNAc coupled to advances in MS technology have drastically increased the number of O-GlcNAc sites that have been identified from the whole proteome of multiple species.28, 62, 79, 80
Figure 5.
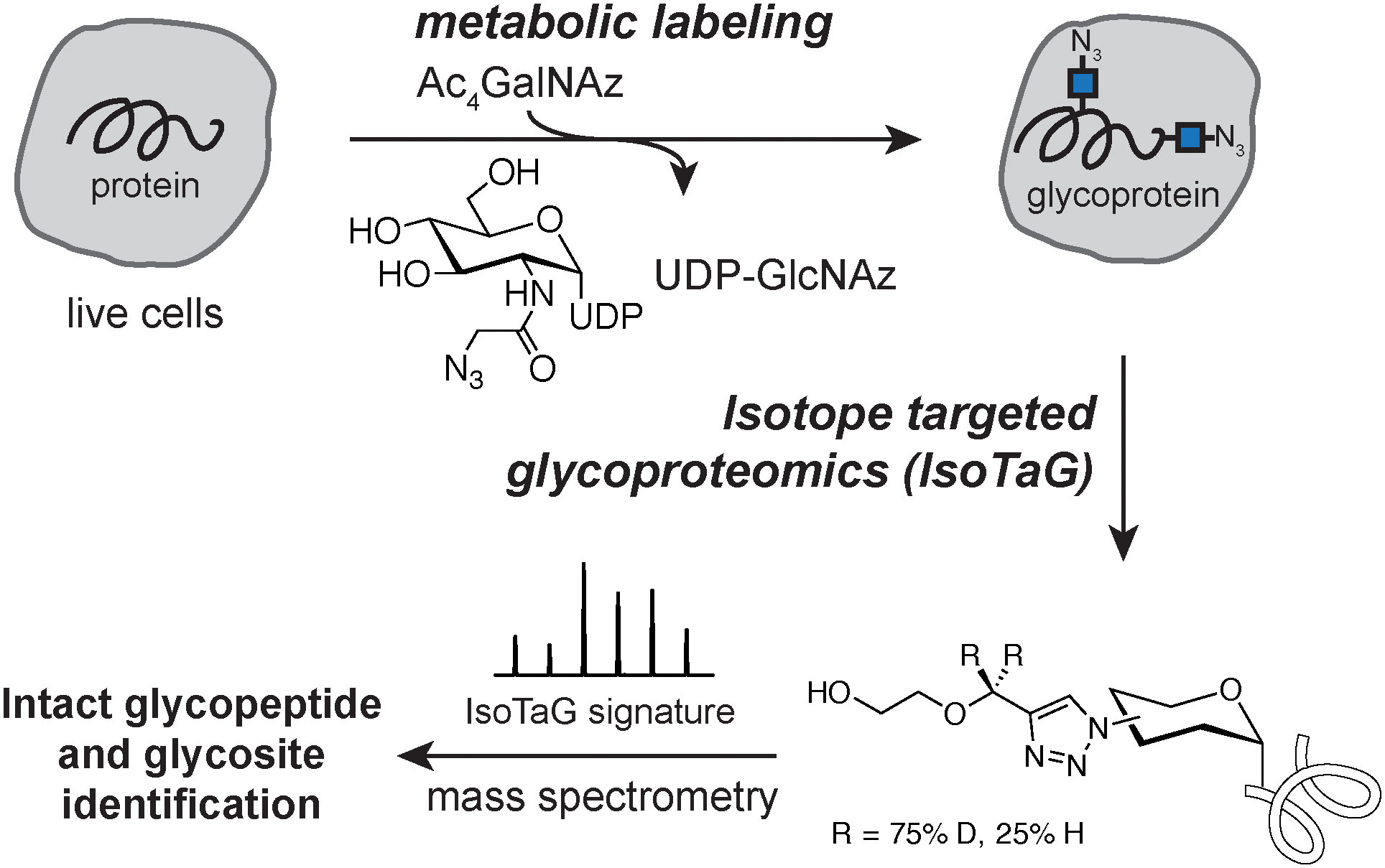
Workflow for Isotope Targeted Glycoproteomics (IsoTaG). Live cells are labeled with an azidosugar (e.g., Ac4GalNAz) as a reporter for the O-GlcNAc modification. Enrichment, digestion, and acid cleavage of the tag recovers the modified glycopeptide for characterization by MS.
Intersection of O-GlcNAc and phosphorylation on the systems-level
The cell integrates glycosylation, phosphorylation, and the myriad of other chemical modifications on a protein into a functional signaling output. Through MS-based proteomics, the ability to map a diversity of modification sites has enhanced significantly over the past decade, concomitantly increasing the depth of maps of the O-GlcNAc proteome.66 In particular, the crosstalk between O-GlcNAc and phosphorylation has long fascinated the field. The possibility of O-GlcNAc blocking proximal phosphosites was first suggested by Hart and coworkers in 1987.18 In 2008, a large-scale phosphosite profiling study in mouse fibroblasts revealed roughly half of the 711 mapped phosphosites changed in abundance in response to a global increase in O-GlcNAc glycosylation via chemical inhibition of OGA.81 Furthermore, inhibition of phosphatase by okadaic acid decreased global O-GlcNAc levels by Western blot in NIH/3T3 cells.82, 83 In primary human T cells, 45% of O-GlcNAc sites occur in close proximity to a previously mapped phosphorylation site (within 10 amino acids).84 However, conflicting evidence for crosstalk between the O-GlcNAc modification and phosphorylation exists. In 2012, clustering of a profile of 1,750 glycosites and 16,500 phosphosites mapped from the murine synaptosome was statistically independent, implying that crosstalk between glycosites and phosphosites at a static state was insignificant.85 In contrast, evidence for reciprocal regulation between O-GlcNAc and phosphorylation was recently demonstrated in vitro on a specific four-amino acid consensus sequence: N-S/T, P, V/A/T, S/T-C.40 Phosphorylation of the sequence at the N-terminal S/T resulted in inhibition of glycosylation at the C-terminal S/T, while glycosylation of the N-terminal S/T resulted in inhibition of phosphorylation at the C-terminal S/T. The observed competitive modification model with this consensus sequence was subsequently evaluated on synthetic peptides derived from ten proteins naturally containing the consensus sequence. All possible permutations of this four-residue sequence are enriched in the human proteome compared to randomly-selected four-residue sequences, potentially indicative of an evolutionary selection for this consensus sequence. Further evaluation of O-GlcNAc and phosphorylation maps from cellular or in vivo systems may reveal instances of PTM crosstalk on a global scale.
O-GlcNAc regulation of kinases
Crosstalk between O-GlcNAc and phosphorylation additionally occurs through the direct O-GlcNAcylation of the kinome, thus regulating downstream phosphorylation events. While crosstalk between O-GlcNAc and phosphorylation is most commonly studied from the perspective of regulation at an individual protein substrate, emerging examples demonstrate that O-GlcNAcylation of the kinase influences substrate selection and enzymatic activity.86, 87 Both OGT and OGA have been immunoprecipitated in protein complexes containing kinases (and phosphatases, see below).88, 89 In vitro glycosylation of a kinase microarray with OGT found that approximately 39% of these kinases are substrates.90 Our analysis of the O-GlcNAc literature and glycoproteomics datasets found over 100 kinases possessing a mapped O-GlcNAc site to date. We review the current O-GlcNAcylated kinome based on large-scale O-GlcNAc maps78, 84, 91–98 and the several examples of the O-GlcNAc modification regulating kinase activity, thereby highlighting the central role of O-GlcNAc in regulating phosphorylation on protein substrates and on the kinases themselves (Figure 6, Table 1).
Figure 6.
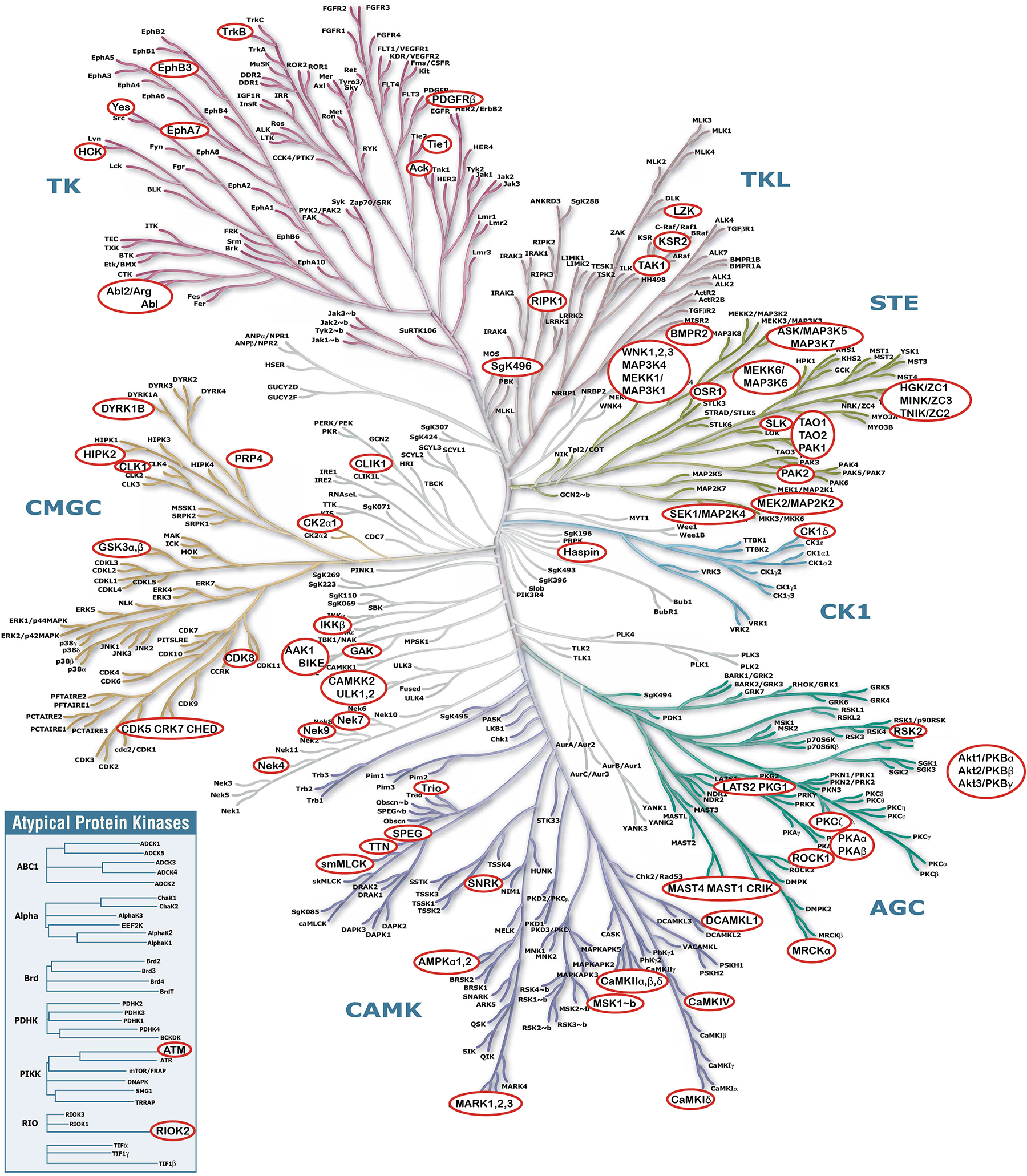
Human kinome with known O-GlcNAc modified kinases circled in red. Illustration reproduced courtesy of Cell Signaling Technology, Inc. (www.cellsignal.com).
Table 1.
Catalog of kinases with at least one mapped glycopeptide reported from biochemical or glycoproteomics studies. For proteins with multiple reported glycosites, sites on different peptides are separated by a comma, while sites found from the same peptide are separated by slashes. If one site contained 3 or more peptide spectral matches (PSMs) across large-scale proteomic studies, that site is bolded. Proteins that had more than five reported glycosites from the same peptide sequence or were ambiguously localized within the peptide from large-scale proteomic studies, are designated as “N/A.”
| Protein names | Gene name(s) | Organism | Reported Glycosite(s) | Uniprot accession(s) | Citation |
|---|---|---|---|---|---|
| 1-phosphatidylinositol 3-phosphate 5-kinase kinase | PIKFYVE, KIAA0981, PIP5K3 | Homo sapiens | T90, S1433/T1436/T1438 | Q9Y2I7 | 84 |
| Activated CDC42 kinase 1 | TNK2 ACK1, Tnk2, Ack1 | Homo sapiens; Mus musculus | N/A | Q07912; O54967 | 92, 30, 91, 93 |
| AP2-associated protein kinase 1 | AAK1, KIAA1048, Aak1, Kiaa1048 | Homo sapiens; Mus musculus | T441/S447/T448, T507/S519; N/A | Q2M2I8; Q3UHJ0 | 92, 30, 84 |
| 6-phosphofructokinase type C, platelet type | PFKP, PFKF | Homo sapiens | T538/S546 | Q01813 | 84 |
| BDNF/NT-3 growth factors receptor | NTRK2 TRKB | Homo sapiens | N/A | Q16620 | 30 |
| BMP-2-inducible protein kinase | BMP2K BIKE HRIHFB2017 | Homo sapiens | T381 | Q9NSY1 | 92, 30 |
| Bone morphogenetic protein receptor type-2 | BMPR2 PPH1, Bmpr2 | Homo sapiens; Mus musculus | T803/T805; S547/S548/S549/ S550 | Q13873; O35607 | 30, 91 |
| Breakpoint cluster region protein | BCR BCR1 D22S11, Bcr Kiaa3017 | Homo sapiens; Mus musculus | S400; N/A | P11274; Q6PAJ1 | 30 |
| Calcium/calmodulin-dependent (CaM) protein kinase I delta, II alpha, II beta, II delta, IV | CAMK1D CAMKID, Camk1d; CAMK2A CAMKA KIAA0968, Camk2a; Camk2b, mCG_122182; Camk2d, Kiaa4163; Camk4 | Homo sapiens; Mus musculus; Caenorhabditis elegans | Human CaMKIV: T57/S58, S137, S189, S344/345, S356 | Q8IU85, Q8BW96; Q9UQM7, P11798; P08414, Q5SVJ0; Q6PHZ2 | 30, 93, 91 |
| Calcium/calmodulin-dependent protein kinase kinase 2 | CAMKK2 CAMKKB KIAA0787 | Homo sapiens | T56 | Q96RR4 | 30 |
| Casein kinase I isoform delta | Csnk1d Hckid | Mus musculus | T344/T347, S382 | Q9DC28 | 91 |
| Casein kinase II subunit alpha | CSNK2A1 CK2A1, Csnk2a1 Ckiia | Homo sapiens; Mus musculus | S347; N/A | P68400; Q60737 | 30, 91 |
| cGMP-dependent protein kinase 1 | PKG1 PRKG1 PRKG1B PRKGR1A PRKGR1B | Homo sapiens | T151 | Q13976 | 84 |
| Citron Rho-interacting kinase | CIT CRIK KIAA0949 STK21 | Homo sapiens | N/A | O14578 | 30 |
| Creatine kinase S-type, mitochondrial | Ckmt2 | Rattus norvegicus | S51 | A0A0G2JVQ1 | 122 |
| Cyclin-dependent kinase 8, 12, 13 | CDK8, Cdk8; CDK12 CRK7 CRKRS KIAA0904, Cdk12 Crk7 Crkrs Kiaa0904; CDK13 CDC2L CDC2L5 CHED KIAA1791, Cdk13 Cdc2l5 Kiaa1791 | Homo sapiens, Mus musculus | Human CDK12: T592/S593/S597/ S601 Human CDK13: T1286/S1287/T1292 Mouse CDK13: T1286 |
P49336, Q8R3L8; Q9NYV4, Q14AX6; Q14004, Q69ZA1 | 96, 97, 98, 92, 84, 91 |
| Cyclin-G-associated kinase | GAK | Homo sapiens | S1130 | O14976 | 84 |
| DDB1- and CUL4-associated factor 1 | DCAF1 KIAA0800 RIP VPRBP | Homo sapiens | T891 | Q9Y4B6 | 30 |
| Diacylglycerol kinase | Dgkh, Dgkd | Mus musculus | N/A | D3YXJ0, E9PUQ8 | 91 |
| Dual serine/threonine and tyrosine protein kinase | DSTYK KIAA0472 RIP5 RIPK5 SGK496 HDCMD38P | Homo sapiens | N/A | Q6XUX3 | 92 |
| Dual specificity mitogen-activated protein kinase kinase 2, 4 | MAP2K2 MEK2 MKK2 PRKMK2; MAP2K4 JNKK1 MEK4 MKK4 PRKMK4 SEK1 SERK1 SKK1 | Homo sapiens | T396/T398; S7/S36 | P36507; P45985 | 84 |
| Dual specificity protein kinase CLK1 | CLK1 CLK | Homo sapiens | T302 | P49759 | 84 |
| Dual specificity tyrosine-phosphorylation-regulated kinase 1B | Dyrk1b | Mus musculus | S624 | Q9Z188 | 91 |
| Ephrin type-A receptor 7 | EPHA7 EHK3 HEK11 | Homo sapiens | T437/S444 | Q15375 | 84 |
| Ephrin type-B receptor 3 | EPHB3 ETK2 HEK2 TYRO6 | Homo sapiens | S390 | P54753 | 84 |
| Glycogen synthase kinase-3 alpha, beta | GSK3A; Gsk3B | Homo sapiens; Mus musculus | T106; T7/T8/S9 | P49840; Q9WV60 | 30, 91 |
| Homeodomain-interacting protein kinase 2 | HIPK2, Hipk2 Nbak1 Stank | Homo sapiens; Mus musculus | N/A; S1009 | Q9H2X6; Q9QZR5 | 98, 93 |
| Inhibitor of nuclear factor kappa-B kinase subunit beta | IKBKB, IKKB | Homo sapiens; Mus musculus | N/A | O14920; O88351 | 123 |
| Kinase superfamily with octicosapeptide/Phox/Bem1p domain-containing protein | At2g35050, At1g79570, At5g57610 | Arabidopsis thaliana | T13 | O64768, A0A1P8ASK1, Q9FKL3 | 91, 94 |
| Kinase suppressor of Ras 2 | Ksr2 | Mus musculus | T212 | Q3UVC0 | 91 |
| MAP/microtubule affinity-regulating kinase 3 | Mark3 Emk2 Mpk10 | Mus musculus | S494/S495 | Q03141 | 91 |
| Microtubule-associated serine/threonine-protein kinase 1, 4 | MAST1 KIAA0973 SAST; Mast4 | Homo sapiens; Mus musculus | S799; S2165 | Q9Y2H9; Q811L6 | 84, 91, 93 |
| Misshapen-like kinase 1 | MINK1 B55 MAP4K6 MINK YSK2 ZC3, Mink1 Map4k6 Mink | Homo sapiens; Mus musculus | T672; T675/S676/S677/ S685 | Q8N4C8; Q9JM52 | 30, 91 |
| Mitogen-activated protein kinase 18 | MPK18 At1g53510 F22G10.12 T3F20.17 | Arabidopsis thaliana | S415/T416 | Q9C5C0 | 91, 94 |
| Mitogen-activated protein kinase kinase kinase 1, 4, 5, 6, 7, 13 | MAP3K1 MAPKKK1 MEKK MEKK1; MAP3K4 KIAA0213 MAPKKK4 MEKK4 MTK1; Map3k5 Ask1 Mekk5; MAP3K6 ASK2 MAPKKK6 MEKK6; MAP3K7 TAK1, Map3k7 Tak1; MAP3K13 LZK | Homo sapiens; Mus musculus | S297; S201, T300; T1227/S1228; N/A; T446/T448; T440/S441; S492/S495; T560 | Q13233; Q9Y6R4; O35099; O95382; O43318, Q62073, A2AP92; O43283 | 84, 30, 91, 92 |
| Mitogen-activated protein kinase kinase kinase kinase 4 | MAP4K4 HGK ZC1 KIAA0687 NIK | Homo sapiens | T840 | O95819 | 84 |
| Myosin light chain kinase, smooth muscle | MYLK MLCK MLCK1 MYLK1 | Homo sapiens | N/A | Q15746 | 92 |
| Non-specific serine/threonine protein kinase | Akt3 | Mus musculus | N/A | Q6NXW0 | 91 |
| Pantothenate kinase 2, mitochondrial | PANK2 C20orf48 | Homo sapiens | T182 | Q9BZ23 | 84 |
| Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, beta, delta | PIK3C2A; PIK3C2B; PIK3CD | Homo sapiens | T334, T1327/S1335; N/A; S950 | O00443; O00750; O00329 | 30, 78, 84 |
| Phosphoenolpyruvate carboxykinase (ATP)-like protein | At4g37870 | Arabidopsis thaliana | N/A | Q0WWL8 | 91, 94 |
| Phosphoglycerate kinase 1 | PGK1 PGKA MIG10 OK/SW-cl.110 | Homo sapiens | N/A | P00558 | 78 |
| Phosphotransferases/inositol or phosphatidylinositol kinase | At4g36080 | Arabidopsis thaliana | S3248 | F4JPL0 | 91, 94 |
| Platelet-derived growth factor receptor beta | Pdgfrb Pdgfr Pdgfr1 | Mus musculus | S308 | P05622 | 91 |
| Probable gluconokinase | IDNK C9orf103 | Homo sapiens | S125 | Q5T6J7 | 84 |
| Protein kinase superfamily protein | At1g73460 T9L24.35 T9L24_35 | Arabidopsis thaliana | S191/S193 | F4HQ88 | 91, 94 |
| Putative 3-phosphoinositide-dependent protein kinase 2 | PDPK2P PDPK2 | Homo sapiens | T182 | Q6A1A2 | 84 |
| Pyruvate kinase PKM | PKM OIP3 PK2 PK3, Pkm Pk3 Pkm2 Pykm | Homo sapiens; Mus musculus | T50; S37 | P14618; P52480 | 30, 91 |
| Receptor-interacting serine/threonine-protein kinase 1 | RIPK1 RIP RIP1 | Homo sapiens | S330/S331 | Q13546 | 30 |
| Rho-associated protein kinase 1 | ROCK1 | Homo sapiens | S1336 | Q13464 | 84 |
| Ribosomal protein S6 kinase alpha-3 | RPS6KA3 ISPK1 MAPKAPK1B RSK2 | Homo sapiens | S556 | P51812 | 84 |
| Ribosomal protein S6 kinase alpha-5 | RPS6KA5 MSK1 | Homo sapiens | S187 | O75582 | 84 |
| Serine-protein kinase ATM | ATM | Homo sapiens | S601, S941, T2608 | Q13315 | 84 |
| Serine/threonine-protein kinase 35 | STK35 CLIK1 PDIK1 STK35L1 | Homo sapiens | N/A | Q8TDR2 | 92 |
| Serine/threonine-protein kinase DCLK1 | Dclk1 Dcamkl1 Dclk | Mus musculus | T156 | Q9JLM8 | 91 |
| Serine/threonine-protein kinase haspin | HASPIN GSG2 | Homo sapiens | S299 | Q8TF76 | 84 |
| Serine/threonine-protein kinase LATS2 | LATS2 KPM | Homo sapiens | S277 | Q9NRM7 | 84 |
| Serine/threonine-protein kinase LMTK3 | LMTK3 KIAA1883 TYKLM3, Lmtk3 Aatyk3 | Homo sapiens; Mus musculus | S1316; S535, S1280 | Q96Q04; Q5XJV6 | 30, 91 |
| Serine/threonine-protein kinase MARK1 | Mark1 Emk3 Kiaa1477 | Mus musculus | N/A | Q8VHJ5 | 91 |
| Serine/threonine-protein kinase MARK2 | Mark2 Emk | Mus musculus | N/A | Q05512 | 91 |
| Serine/threonine-protein kinase MRCK alpha | CDC42BPA KIAA0451, Cdc42bpa Kiaa0451 | Homo sapiens; Mus musculus | S1609; S1596 | Q5VT25; Q3UU96 | 30, 91 |
| Serine/threonine-protein kinase N1 | PKN1 PAK1 PKN PRK1 PRKCL1 | Homo sapiens | S559/S561 | Q16512 | 84 |
| Serine/threonine-protein kinase Nek4, Nek7, Nek9 | NEK4 STK2; NEK7; NEK9 KIAA1995 NEK8 NERCC | Homo sapiens | Nek7: S260/S264 Nek9: T877 |
P51957; Q8TDX7; Q8TD19 | 92, 84, 30 |
| Serine/threonine-protein kinase OSR1 | OXSR1 KIAA1101 OSR1 | Homo sapiens | N/A | O95747 | 92 |
| Serine/threonine-protein kinase PAK 2 | PAK2 | Homo sapiens | T169/T178 | Q13177 | 84 |
| Serine/threonine-protein kinase PRP4 | PRPF4B KIAA0536 PRP4 PRP4H PRP4K | Homo sapiens | S20, S356 | Q13523 | 84 |
| Serine/threonine-protein kinase RIO2 | RIOK2 RIO2 | Homo sapiens | S239 | Q9BVS4 | 84 |
| Serine/threonine-protein kinase TAO1, TAO2 | TAOK1 KIAA1361 MAP3K16 MARKK; TAOK2 KIAA0881 MAP3K17 PSK PSK1 UNQ2971/PRO7431 | Homo sapiens | N/A | Q7L7X3; Q9UL54 | 92 |
| Serine/threonine-protein kinase ULK2 | ULK2 KIAA0623, Ulk2 Kiaa0623 | Homo sapiens; Mus musculus | T613; T613, T727 | Q8IYT8; Q9QY01 | 30, 91, 93 |
| Serine/threonine-protein kinase WNK1, WNK2, WNK3 | WNK1 HSN2 KDP KIAA0344 PRKWNK1, Wnk1 Hsn2 Prkwnk1; WNK2 KIAA1760 PRKWNK2 SDCCAG43 P/OKcl.13, Wnk2 Kiaa1760; WNK3 KIAA1566 PRKWNK3, Wnk3 | Homo sapiens, Mus musculus | WNK1 Human: T1848/S1849/S1850 WNK1 Mouse: S1230, S1844, T1945, T2291, S2301, T2376 WNK2 Human: T1604/S1606 WNK2 Mouse: T1698 WNK3 Human: T709, T974, T1312 WNK3 Mouse: S1161 |
Q9H4A3, P83741; Q9Y3S1, Q3UH66; Q9BYP7, Q80XP9 | 92, 30, 84; 91, 93 |
| SNF-related serine/threonine-protein kinase | Snrk | Mus musculus | N/A | Q8VDU5 | 91 |
| STE20-like serine/threonine-protein kinase | Slk Kiaa0204 Stk2 | Mus musculus | S1230 | O54988 | 91 |
| Striated muscle preferentially expressed protein kinase | SPEG APEG1 KIAA1297 | Homo sapiens | S1880 | Q15772 | 84 |
| Titin | TTN | Homo sapiens | S4651, T4659, S10385, S33976 | Q8WZ42 | 84 |
| TRAF2 and NCK-interacting protein kinase | TNIK KIAA0551, Tnik Kiaa0551 | Homo sapiens; Mus musculus | S568; S539, T577 | Q9UKE5; P83510 | 92, 30; 93 |
| Triple functional domain protein | Trio | Mus musculus | N/A | Q0KL02 | 91 |
| Tyrosine-protein kinase | Abl2 | Mus musculus | T822 | B2RQ57 | 91 |
| Tyrosine-protein kinase ABL1, ABL2 | ABL1 ABL JTK7; Abl2 Arg | Homo sapiens; Mus musculus | S855; T872 | P00519; Q4JIM5 | 30, 93 |
| Tyrosine-protein kinase HCK | HCK | Homo sapiens | S347/S351 | P08631 | 84 |
| Tyrosine-protein kinase receptor Tie-1 | TIE1 TIE | Homo sapiens | N/A | P35590 | 92 |
| Tyrosine-protein kinase Yes | Yes1 Yes | Mus musculus | N/A | Q04736 | 91 |
| Uridine-cytidine kinase 2 | UCK2 UMPK | Homo sapiens | T106 | Q9BZX2 | 84 |
| Bifunctional UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase | GNE | Homo sapiens | N/A | Q9Y223 | 114 |
| 5’-AMP-activated protein kinase | PRKAA2, PRKACA; PRKACB; AMPK | Homo sapiens | N/A | P54646, P17612, P22694 | 111 99 |
| RAC-alpha serine/threonine-protein kinase 1; 2 | AKT1; AKT2 | Homo sapiens; Rattus norvegicus | T305/T312; N/A | P31749; P47197 | 100, 104 |
| Protein kinase C zeta type | Prkcz | Mus musculus | T410 (also a phosphosite) | Q02956 | 124 |
| Cyclin-dependent-like kinase 5 | CDK5 | Homo sapiens | S46, T245/T246/S247 | Q00535 | 125 |
| Serine/threonine-protein kinase ULK1 | ULK1 KIAA0722 | Homo sapiens | T754 | O75385 | 110 |
AGC
The AGC kinase family contains Ser/Thr protein kinases named after three representative families, the cAMP-dependent protein kinase (PKA), the cGMP-dependent protein kinase (PKG) and the protein kinase C (PKC) families. The AGC family contains more than 60 human protein kinases. To our knowledge, seven members of this kinase family have been identified as O-GlcNAc modified.
PKA plays a role in CREB signaling and additionally modifies tau. O-GlcNAcylation of PKA subunit PKAcα and PKAcβ alters their subcellular localization and enhances their kinase activity.99 Protein kinase B (AKT) is a serine/threonine kinase involved in multiple cellular processes, including the insulin response, apoptosis, and cell migration. AKT was found to be modified by O-GlcNAc in 2006.100 Enrichment of AKT by wheat germ agglutinin increased under high glucose conditions or when dosing the OGA inhibitor PUGNAc, indicative of higher O-GlcNAc levels on AKT. Follow up experiments demonstrated that elevated global O-GlcNAc levels correlate with AKT translocation from the cytoplasm to the nucleus. Two separate laboratories later reported distinct glycosites on AKT.101, 102 Hart and co-workers used tyrosine as a glyco-mimetic mutation to show that O-GlcNAcylation at T305 and/or T312 on AKT inhibits phosphorylation at T308, a residue in the activation loop of AKT whose phosphorylation is necessary for AKT activation.103 These studies implicated glycosylation as a mechanism to downregulate AKT activity using an in vitro AKT activity assay.101 Gong and co-workers later report that increased O-GlcNAcylation on AKT positively correlates with markers for apoptosis and overexpression of AKT alleviates this phenotype.102 AKT2 has also been identified as O-GlcNAc modified.104 Treatment of rat adipocytes with the OGA inhibitor PUGNAc increased glycosylation and decreased insulin-induced phosphorylation of AKT2 by Western blot.
O-GlcNAc may be linked to regulation of cancer cell migration via another member of the AGC kinase family, ROCK.105 Chemical inhibition of OGA resulted in accelerated migration that was found to be mediated by the RhoA/ROCK/MLC (myosin light chain) interaction in SKOV3 and 59M ovarian cancer cells. Knockout RhoA or inhibition of ROCK eliminates the change in cell migration caused by OGA inhibition. While direct O-GlcNAcylation of RhoA or ROCK was not established in this study, a glycosite on ROCK has recently been identified in a large-scale glycoproteomics study.84 Biochemical confirmation and characterization of this glycosite will lead to further conclusions about the role of O-GlcNAc in regulating ROCK and cell migration.
CMGC
The CMGC family of kinases contains the cyclin-dependent kinases (CDK), mitogen-activated protein kinases (MAPK), glycogen synthase kinases, and the CDC-like kinases (CLK). These kinases are involved in cell-cycle regulation and signaling, cell communication, and cell growth.
GSK3β regulates OGT via phosphorylation and is likewise regulated by OGT through O-GlcNAc. Inhibition of GSK3β alters the abundance of several O-GlcNAc sites on GSK3β in the mouse and monkey proteomes, a result that could be attributed to the loss of phosphorylation on OGT or other GSK3β substrates.106, 107 In addition, the O-GlcNAc modification on GSK3β results in regulation of the molecular chaperones that are stably expressed under heat-shock conditions.108 Specifically, knockout of OGT in MEF cells results in altered expression of HSP72, a heat shock protein that is governed by the GSK3β-substrate HSF1.
Several studies additionally report regulation of CMGC complexes by O-GlcNAc. In these examples, members of the CMGC kinase complex are glycosylated, leading to alteration of the kinase function and substrate selectivity that affect essential cellular processes, including motility and cell cycle progression. For example, p27 is a tumor suppressor gene that functions by inhibiting CDK2/Cyclin E. Shen and co-workers mapped glycosites on p27 at Ser2, Ser106, Ser110, Thr157, and Thr198.109 O-GlcNAcylation of Ser2 destabilizes p27 and works synergistically with phosphorylation of Ser10 to move the cell cycle forward. Furthermore, Western blot and immunohistochemical analyses of hepatocellular carcinoma tissues and their corresponding nontumorous tissues were performed, and revealed that elevated O-GlcNAc on p27 correlates with increased cell proliferation. Together, these results indicate that the dynamic interplay between O-GlcNAcylation and phosphorylation on p27 in complex with CDK2 controls CDK2 activity via regulation of p27 stability.
CAMK
The calcium/calmodulin-dependent kinases (CAMK) are a family of enzymes stimulated by calmodulin, a protein that is activated in response to increased intracellular calcium concentrations. As many CAMK substrates are transcription factors, this kinase family is known for being closely tied to regulation of gene expression. A number of CAMK subunits are modified by OGT, with the effects on CAMKIV being best studied.86 CAMKIV is O-GlcNAcylated at several sites. Modification of CAMKIV with O-GlcNAc at the active site reduces the level of stimulatory phosphorylation at T200 and results in inhibition of the kinase activity.
Unc-51-like-kinase 1 (ULK1) is an important gatekeeper of the autophagy pathway. ULK1 is glycosylated at T754. Glycosylation of ULK1 can only occur once ULK1 has been dephosphorylated by PP1 to remove a phosphosite installed by mTOR.110 O-GlcNAcylation of ULK1 at T754 promotes binding to substrate ATG14L, which results in phosphatidylinositol-(3)-phosphate production and initiation of autophagy. In this example, dephosphorylation by PP1 represents a gatekeeping step for subsequent O-GlcNAcylation, and illustrates key regulatory mechanisms by O-GlcNAc in the autophagy pathway.
AMPK is a heterotrimeric kinase that has a protective function from cellular metabolic stress. AMPK activity is strongly associated with depleted cellular energy levels as the kinase is activated by 5’-AMP and ADP, but inhibited by ATP. Stimulation with 5’-AMP or ADP yields a net upregulation of catabolic and downregulation of anabolic processes. The kinase complex is further activated by phosphorylation of T172 in the AMPK alpha subunit. During differentiation of C2C12 mouse skeletal muscle myotubes, AMPK activity is closely associated with OGT translocation to the nucleus. The altered localization of OGT results in increased O-GlcNAcylation of nuclear proteins and H3K9 acetylation111 and results in phosphorylation of OGT at T444. Phosphorylation of OGT by AMPK alters the O-GlcNAc landscape. All α and γ subunits of AMPK substrates for OGT, and active AMPK shows increased O-GlcNAcylation of the γ1 subunit.
STE
The homologs of yeast Sterile 7, 11, and 20 kinase family contains many kinases involved in cell growth, differentiation, oxidative damage, and apoptosis, including many MAP and serine/threonine protein kinases.112, 113 PAK, GCK, MEK, and MKK kinases are also part of the STE family. Prominent O-GlcNAcylated members of this family include WNK1–3, PAK1–2, TAO1–2, SLK, OSR1, and several MAP3K proteins, all revealed in large-scale proteomics experiments. The functional outcomes of glycosites on kinases in the STE family have yet to be biochemically characterized.
CK1
Despite the limited number of kinases in the CK1 family, kinases in the CK1 family are involved in regulation of membrane transport, cell division, DNA repair, and nuclear localization. CK1δ and CK2α have been discovered to be O-GlcNAc modified to date. The O-GlcNAcylation of CK1δ was discovered in a large-scale murine synaptosome proteomics study at multiple potential sites that remain to be functionally characterized.91 Glycosylation of human CK2α was evaluated by Cole and coworkers.87 The mapped glycosite S347 was found proximal to multiple known phosphosites on CK2α. By semi-synthesis, O-GlcNAcylation of CK2α was found to inhibit phosphorylation at T344, which decreased the interaction of CK2α with Pin1 and produced a net destabilization of CK2α.
TK/TKL
The protein tyrosine kinase family can be subdivided into two main groups: cytosolic tyrosine kinases (CTKs) (e.g., Src, JAK, Abl) and receptor tyrosine kinases (RTKs) (e.g., EGFR, VEGFR, FLT3). Receptor tyrosine kinases are transmembrane proteins that are activated by the binding of an extracellular ligand that induces dimerization and subsequent autophosphorylation of two RTK monomers, followed by phosphorylation of downstream signaling proteins. Since tyrosine kinases regulate many key processes including cell growth and survival, their dysregulation has been found in the development and progression of a wide range of cancers. The tyrosine kinase-like family (TKL) is closely related to TK, but its members are serine/threonine kinases instead (e.g. Raf). Diverse members of this kinase family are shown to be glycosylated, including those involved in cell differentiation (BMPR2).30, 84, 91 Regulatory functions for the O-GlcNAc sites on kinases in this family await biochemical characterization.
Other kinases
Kinases that phosphorylate non-protein targets are also privy to regulation by O-GlcNAc. GNE is an epimerase/kinase responsible for converting UDP-GlcNAc to ManNAc-6P.114 A GNE point mutation at M743T, commonly observed in GNE myopathy, results in significantly higher O-GlcNAcylation of GNE than its wildtype counterpart. Elevated O-GlcNAcylation on GNE was found to inhibit the epimerase activity of both the wildtype enzyme and the M743T mutant. One hypothesis for why GNE is regulated by O-GlcNAc is due to its role in the consumption of UDP-GlcNAc, the donor sugar used by OGT from the hexosamine biosynthesis pathway.
OGT modifies kinases involved in sugar metabolism and glycolysis in addition to GNE, including pyruvate kinase M2 and phosphofructokinase 1.115, 116 Phosphofructokinase 1 (PFK1) is an enzyme in glycolysis responsible for converting fructose-6-phosphate to fructose-1,6-bisphosphate, a committal step that sends the product through the rest of the glycolytic pathway as compared to the hexosamine biosynthetic pathway. OGT installs O-GlcNAc at S529 of PFK1, attenuating its kinase activity.116 This modification increases in abundance under hypoxic conditions, leading to the redirection of glycolytic flux from the glycolysis pathway toward the pentose phosphate pathway, a glucose-consuming metabolic pathway necessary for synthesis of nucleotides and other sugars.
Phosphatases
Protein phosphatases are a family of approximately 200 enzymes that remove phosphorylation from the protein substrate. Several examples of interplay between O-GlcNAc and dephosphorylation have been reported, including the identification of a functional complex between OGT and protein phosphatase 1.117 For example, priming phosphorylation of folliculin-interacting protein 1 (FNIP1) at S938 by CK2 leads to many subsequent phosphorylation events of FNIP1, ultimately resulting in binding to Hsp90 to inhibit its ATPase activity. If this priming phosphorylation does not occur, OGT can glycosylate FNIP1 at S938, blocking subsequent phosphorylation steps, and consequently lead FNIP1 to be ubiquitinated and degraded.118 Activation of the transcription factor Sp1 is enhanced on dephosphorylation by phosphatase 2A and may be additionally controlled by reciprocal O-GlcNAc and phosphate modification.119
A number of phosphatases are also privy to modification by O-GlcNAc (e.g., MYPT1, PPFIA2–4, PPP6R2, PTPN6, PTPN7, PTPRC, TNS2, SIRPA).84, 85, 92, 93 Direct regulation of phosphatase activity by O-GlcNAc has been reported in a few instances. O-GlcNAcylation of protein tyrosine phosphatase 1B (PTP1B) at S104, S201, and S386 inhibits PTP1B activity, which leads to an increase in AKT and GSK3β activity and therefore insulin response in HepG2 cells.120 Human small CTD phosphatase 1 (hSCP1) was identified as O-GlcNAc modified by Western blot, and its glycosite at S41 was confirmed by Q-TOF MS and site-directed mutagenesis.121 Additionally, the phosphatase myosin phosphatase target subunit 1 (MYPT1) may regulate the substrate specificity of OGT.89 MYPT1 and OGT can be co-immunoprecipitated, MYPT1 is modified by O-GlcNAc, and depletion of MYPT1 alters OGT substrate selectivity in Neuro-2a neuroblastoma cells. These studies highlight additional mechanisms of cellular integration of the O-GlcNAc modification and phosphorylation signaling and a significant opportunity for further study.
Conclusion
The O-GlcNAc modification has emerged a prominent regulator of phosphorylation during cellular signaling via tuning kinase activity in addition to crosstalk between O-GlcNAc and phosphorylation on protein substrates. Regulation of kinases by the O-GlcNAc modification may enable cells to manage resources in disparate pathways according to nutrient availability, and thus finely tune signaling pathways through other modifications like phosphorylation. With the convergence of methods to study and engineer O-GlcNAc on the systems scale and on individual proteins emerging, the increasing evaluation of the functions for O-GlcNAc on kinases and the enzymes that install it will yield a wealth of insights to regulatory mechanisms cells use to integrate these pathways. In particular, the role of O-GlcNAcylation in the STE, TK, and TKL kinase families awaits elucidation. Due to the global nature of O-GlcNAc in cells, further illumination of the functions of O-GlcNAc on kinases will lead to important discoveries in cellular regulation and dysregulation relevant to all areas of biology under normal physiology or disease.
Keywords.
O-linked N-acetyl glucosamine (O-GlcNAc): a carbohydrate that is installed on serine/threonine residues of nuclear/cytosolic proteins by O-GlcNAc transferase (OGT) and removed by O-GlcNAcase (OGA)
Glycosylation: the enzymatic addition of a sugar molecule to another biomolecule
Kinase: an enzyme that catalyzes covalent attachment of a phosphate group to its substrate
Phosphatase: an enzyme that catalyzes the removal of a phosphate group from its substrate
Phosphorylation: the enzymatic addition of a phosphate group to another molecule
Glycoproteomics: the identification and characterization of carbohydrate-modified proteins from a biological sample in the whole proteome via a profiling method (e.g., mass spectrometry)
Crosstalk: the phenomenon where changes in one biological pathway directly affects signaling in another biological pathway
Post-Translational Modification (PTM): the chemical modification of proteins after protein biosynthesis, often catalyzed by enzymes
PTM Crosstalk: the presence of one PTM affecting the substitution pattern of another PTM
Signaling: the transduction of a signal via non-covalent or covalent associations of biological molecules within a pathway
Acknowledgements
We thank B. Yang and C. Aonbangkhen for helpful discussions. Support from the National Institutes of Health (U01CA242098-01, C.M.W.), Burroughs Welcome Fund, Career Award at the Scientific Interface (C.M.W.), Sloan Foundation (C.M.W.), and Harvard University is gratefully acknowledged.
References
- [1].Torres CR, and Hart GW (1984) Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc, J Biol Chem 259, 3308–3317. [PubMed] [Google Scholar]
- [2].Zachara N, Akimoto Y, and Hart GW (2015) The O-GlcNAc Modification, In Essentials of Glycobiology (Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, Schnaar RL, and Seeberger PH, Eds.), pp 239–251, Cold Spring Harbor Laboratory Press, Cold Spring Harbor (NY). [Google Scholar]
- [3].Hardiville S, and Hart GW (2014) Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation, Cell Metab 20, 208–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Olszewski NE, West CM, Sassi SO, and Hartweck LM (2010) O-GlcNAc protein modification in plants: Evolution and function, Biochim Biophys Acta 1800, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Machida M, and Jigami Y (1994) Glycosylated DNA-Binding Proteins from Filamentous Fungus, Aspergillus-Oryzae - Modification with N-Acetylglucosamine Monosaccharide through an O-Glycosidic Linkage, Biosci Biotechn Bioch 58, 344–348. [Google Scholar]
- [6].Ostrowski A, Gundogdu M, Ferenbach AT, Lebedev AA, and van Aalten DM (2015) Evidence for a Functional O-Linked N-Acetylglucosamine (O-GlcNAc) System in the Thermophilic Bacterium Thermobaculum terrenum, J Biol Chem 290, 30291–30305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zentella R, Sui N, Barnhill B, Hsieh W-P, Hu J, Shabanowitz J, Boyce M, Olszewski NE, Zhou P, Hunt DF, and Sun T. p. (2017) The Arabidopsis O-fucosyltransferase SPINDLY activates nuclear growth repressor DELLA, Nat Chem Biol 13, 479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Halim A, Larsen IS, Neubert P, Joshi HJ, Petersen BL, Vakhrushev SY, Strahl S, and Clausen H (2015) Discovery of a nucleocytoplasmic O-mannose glycoproteome in yeast, P Natl Acad Sci U S A 112, 15648–15653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zachara NE, and Hart GW (2004) O-GlcNAc a sensor of cellular state: the role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress, Biochim Biophys Acta 1673, 13–28. [DOI] [PubMed] [Google Scholar]
- [10].Hart GW, Slawson C, Ramirez-Correa G, and Lagerlof O (2011) Cross Talk Between O-GlcNAcylation and Phosphorylation: Roles in Signaling, Transcription, and Chronic Disease, In Ann Rev Biochem (Kornberg RD, Raetz CRH, Rothman JE, and Thorner JW, Eds.), pp 825–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hanover JA (2001) Glycan-dependent signaling: O-linked N-acetylglucosamine, FASEB J 15, 1865–1876. [DOI] [PubMed] [Google Scholar]
- [12].Dias WB, and Hart GW (2007) O-GlcNAc modification in diabetes and Alzheimer’s disease, Mol Biosyst 3, 766–772. [DOI] [PubMed] [Google Scholar]
- [13].Comer FI, and Hart GW (2000) O-Glycosylation of nuclear and cytosolic proteins. Dynamic interplay between O-GlcNAc and O-phosphate, J Biol Chem 275, 29179–29182. [DOI] [PubMed] [Google Scholar]
- [14].Hart GW, Housley MP, and Slawson C (2007) Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins, Nature 446, 1017–1022. [DOI] [PubMed] [Google Scholar]
- [15].Bond MR, and Hanover JA (2013) O-GlcNAc cycling: a link between metabolism and chronic disease, Annu Rev Nutr 33, 205–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zachara NE, and Hart GW (2006) Cell signaling, the essential role of O-GlcNAc!, Biochim Biophys Acta 1761, 599–617. [DOI] [PubMed] [Google Scholar]
- [17].Slawson C, and Hart GW (2011) O-GlcNAc signalling: implications for cancer cell biology, Nat Rev Cancer 11, 678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Holt GD, Haltiwanger RS, Torres CR, and Hart GW (1987) Erythrocytes contain cytoplasmic glycoproteins. O-linked GlcNAc on Band 4.1, J Biol Chem 262, 14847–14850. [PubMed] [Google Scholar]
- [19].Walsh C (2006) Posttranslational modification of proteins: expanding nature’s inventory, Roberts and Company Publishers. [Google Scholar]
- [20].Pravata VM, Muha V, Gundogdu M, Ferenbach AT, Kakade PS, Vandadi V, Wilmes AC, Borodkin VS, Joss S, Stavridis MP, and van Aalten DMF (2019) Catalytic deficiency of O-GlcNAc transferase leads to X-linked intellectual disability, P Natl Acad Sci U S A 116, 14961–14970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shafi R, Iyer SP, Ellies LG, O’Donnell N, Marek KW, Chui D, Hart GW, and Marth JD (2000) The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny, P Natl Acad Sci 97, 5735–5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yang YR, Song M, Lee H, Jeon Y, Choi EJ, Jang HJ, Moon HY, Byun HY, Kim EK, Kim DH, Lee MN, Koh A, Ghim J, Choi JH, Lee-Kwon W, Kim KT, Ryu SH, and Suh PG (2012) O-GlcNAcase is essential for embryonic development and maintenance of genomic stability, Aging Cell 11, 439–448. [DOI] [PubMed] [Google Scholar]
- [23].O’Donnell N, Zachara NE, Hart GW, and Marth JD (2004) Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability, Mol Cell Biol 24, 1680–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Levine ZG, and Walker S (2016) The Biochemistry of O-GlcNAc Transferase: Which Functions Make It Essential in Mammalian Cells?, Ann Rev Biochem 85, 631–657. [DOI] [PubMed] [Google Scholar]
- [25].Lagerlöf O, Slocomb JE, Hong I, Aponte Y, Blackshaw S, Hart GW, and Huganir RL (2016) The nutrient sensor OGT in PVN neurons regulates feeding, Science 351, 1293–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Itkonen HM, Minner S, Guldvik IJ, Sandmann MJ, Tsourlakis MC, Berge V, Svindland A, Schlomm T, and Mills IG (2013) O-GlcNAc transferase integrates metabolic pathways to regulate the stability of c-MYC in human prostate cancer cells, Cancer Res 73, 5277–5287. [DOI] [PubMed] [Google Scholar]
- [27].Huang X, Pan Q, Sun D, Chen W, Shen A, Huang M, Ding J, and Geng M (2013) O-GlcNAcylation of cofilin promotes breast cancer cell invasion, J Biol Chem 288, 36418–36425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lund PJ, Elias JE, and Davis MM (2016) Global Analysis of O-GlcNAc Glycoproteins in Activated Human T Cells, J Immunol 197, 3086–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Marotta NP, Lin YH, Lewis YE, Ambroso MR, Zaro BW, Roth MT, Arnold DB, Langen R, and Pratt MR (2015) O-GlcNAc modification blocks the aggregation and toxicity of the protein α-synuclein associated with Parkinson’s disease, Nat Chem 7, 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Liu F, Shi J, Tanimukai H, Gu J, Gu J, Grundke-Iqbal I, Iqbal K, and Gong CX (2009) Reduced O-GlcNAcylation links lower brain glucose metabolism and tau pathology in Alzheimer’s disease, Brain 132, 1820–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Brown JR, Fuster MM, Li R, Varki N, Glass CA, and Esko JD (2006) A disaccharide-based inhibitor of glycosylation attenuates metastatic tumor cell dissemination, Clin Cancer Res 12, 2894–2901. [DOI] [PubMed] [Google Scholar]
- [32].Ramirez DH, Aonbangkhen C, Wu H-Y, Naftaly JA, Tang S, O’Meara TR, and Woo CM (2020) Engineering a proximity-directed O-GlcNAc transferase for selective protein O-GlcNAcylation in cells, ACS Chem Biol, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yang WH, Kim JE, Nam HW, Ju JW, Kim HS, Kim YS, and Cho JW (2006) Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability, Nat Cell Biol 8, 1074–1083. [DOI] [PubMed] [Google Scholar]
- [34].Chou TY, Hart GW, and Dang CV (1995) c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas, J Biol Chem 270, 18961–18965. [DOI] [PubMed] [Google Scholar]
- [35].Yuzwa SA, Shan X, Macauley MS, Clark T, Skorobogatko Y, Vosseller K, and Vocadlo DJ (2012) Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation, Nat Chem Biol 8, 393–399. [DOI] [PubMed] [Google Scholar]
- [36].Brister MA, Pandey AK, Bielska AA, and Zondlo NJ (2014) OGlcNAcylation and phosphorylation have opposing structural effects in tau: phosphothreonine induces particular conformational order, J Am Chem Soc 136, 3803–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chen YX, Du JT, Zhou LX, Liu XH, Zhao YF, Nakanishi H, and Li YM (2006) Alternative O-GlcNAcylation/O-phosphorylation of Ser16 induce different conformational disturbances to the N terminus of murine estrogen receptor beta, Chem Biol 13, 937–944. [DOI] [PubMed] [Google Scholar]
- [38].Mao X, Zhang D, Tao T, Liu X, Sun X, Wang Y, and Shen A (2015) O-GlcNAc glycosylation of p27(kip1) promotes astrocyte migration and functional recovery after spinal cord contusion, Exp Cell Res 339, 197–205. [DOI] [PubMed] [Google Scholar]
- [39].Toleman CA, Schumacher MA, Yu SH, Zeng W, Cox NJ, Smith TJ, Soderblom EJ, Wands AM, Kohler JJ, and Boyce M (2018) Structural basis of O-GlcNAc recognition by mammalian 14-3-3 proteins, P Natl Acad Sci U S A 115, 5956–5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Leney AC, El Atmioui D, Wu W, Ovaa H, and Heck AJR (2017) Elucidating crosstalk mechanisms between phosphorylation and O-GlcNAcylation, P Natl Acad Sci U S A 114, E7255–E7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zeidan Q, and Hart GW (2010) The intersections between O-GlcNAcylation and phosphorylation: implications for multiple signaling pathways, J Cell Sci 123, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Iyer SPN, and Hart GW (2003) Roles of the Tetratricopeptide Repeat Domain in O-GlcNAc Transferase Targeting and Protein Substrate Specificity, J Biol Chem 278, 24608–24616. [DOI] [PubMed] [Google Scholar]
- [43].Lazarus MB, Nam Y, Jiang J, Sliz P, and Walker S (2011) Structure of human O-GlcNAc transferase and its complex with a peptide substrate, Nature 469, 564–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Joiner CM, Levine ZG, Aonbangkhen C, Woo CM, and Walker S (2019) Aspartate Residues Far from the Active Site Drive O-GlcNAc Transferase Substrate Selection, J Am Chem Soc 141, 12974–12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Levine ZG, Fan C, Melicher MS, Orman M, Benjamin T, and Walker S (2018) O-GlcNAc Transferase Recognizes Protein Substrates Using an Asparagine Ladder in the Tetratricopeptide Repeat (TPR) Superhelix, J Am Chem Soc 140, 3510–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lubas WA, and Hanover JA (2000) Functional Expression of O-linked GlcNAc Transferase: DOMAIN STRUCTURE AND SUBSTRATE SPECIFICITY, J Biol Chem 275, 10983–10988. [DOI] [PubMed] [Google Scholar]
- [47].Maynard JC, Burlingame AL, and Medzihradszky KF (2016) Cysteine S-linked N-acetylglucosamine (S-GlcNAcylation), a new post-translational modification in mammals, Mol Cell Proteomics, mcp.M116.061549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Darabedian N, Gao J, Chuh KN, Woo CM, and Pratt MR (2018) The Metabolic Chemical Reporter 6-Azido-6-deoxy-glucose Further Reveals the Substrate Promiscuity of O-GlcNAc Transferase and Catalyzes the Discovery of Intracellular Protein Modification by O-Glucose, J Am Chem Soc 140, 7092–7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lazarus MB, Jiang J, Kapuria V, Bhuiyan T, Janetzko J, Zandberg WF, Vocadlo DJ, Herr W, and Walker S (2013) HCF-1 is cleaved in the active site of O-GlcNAc transferase, Science 342, 1235–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Janetzko J, and Walker S (2017) Aspartate Glycosylation Triggers Isomerization to Isoaspartate, J Am Chem Soc 139, 3332–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kreppel LK, and Hart GW (1999) Regulation of a Cytosolic and Nuclear O-GlcNAc Transferase: ROLE OF THE TETRATRICOPEPTIDE REPEATS, J Biol Chem 274, 32015–32022. [DOI] [PubMed] [Google Scholar]
- [52].Wrabl JO, and Grishin NV (2001) Homology between O-linked GlcNAc transferases and proteins of the glycogen phosphorylase superfamily, J Mol Biol 314, 365–374. [DOI] [PubMed] [Google Scholar]
- [53].Nagel AK, and Ball LE (2014) O-GlcNAc transferase and O-GlcNAcase: achieving target substrate specificity, Amino Acids 46, 2305–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Dong DL, and Hart GW (1994) Purification and characterization of an O-GlcNAc selective N-acetyl-beta-D-glucosaminidase from rat spleen cytosol, J Biol Chem 269, 19321–19330. [PubMed] [Google Scholar]
- [55].Whisenhunt TR, Yang X, Bowe DB, Paterson AJ, Van Tine BA, and Kudlow JE (2006) Disrupting the enzyme complex regulating O-GlcNAcylation blocks signaling and development, Glycobiology 16, 551–563. [DOI] [PubMed] [Google Scholar]
- [56].Alonso J, Schimpl M, and van Aalten DM (2014) O-GlcNAcase: promiscuous hexosaminidase or key regulator of O-GlcNAc signaling?, J Biol Chem 289, 34433–34439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Keembiyehetty CN, Krzeslak A, Love DC, and Hanover JA (2011) A lipid-droplet-targeted O-GlcNAcase isoform is a key regulator of the proteasome, J Cell Sci 124, 2851–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Elsen NL, Patel SB, Ford RE, Hall DL, Hess F, Kandula H, Kornienko M, Reid J, Selnick H, Shipman JM, Sharma S, Lumb KJ, Soisson SM, and Klein DJ (2017) Insights into activity and inhibition from the crystal structure of human O-GlcNAcase, Nat Chem Biol 13, 613–615. [DOI] [PubMed] [Google Scholar]
- [59].Li B, Li H, Lu L, and Jiang J (2017) Structures of human O-GlcNAcase and its complexes reveal a new substrate recognition mode, Nat Struct Mol Biol 24, 362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Roth C, Chan S, Offen WA, Hemsworth GR, Willems LI, King DT, Varghese V, Britton R, Vocadlo DJ, and Davies GJ (2017) Structural and functional insight into human O-GlcNAcase, Nat Chem Biol 13, 610–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zhang Z, Tan EP, VandenHull NJ, Peterson KR, Slawson C (2014) O-GlcNAcase expression is sensitive to changes in O-GlcNAc homeostasis, Front Endocrinol 5, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Khidekel N, Ficarro SB, Clark PM, Bryan MC, Swaney DL, Rexach JE, Sun YE, Coon JJ, Peters EC, and Hsieh-Wilson LC (2007) Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics, Nat Chem Biol 3, 339–348. [DOI] [PubMed] [Google Scholar]
- [63].Woo CM, Iavarone AT, Spiciarich DR, Palaniappan KK, and Bertozzi CR (2015) Isotope-targeted glycoproteomics (IsoTaG): a mass-independent platform for intact N- and O-glycopeptide discovery and analysis, Nat Meth 12, 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Nilsson J, Halim A, Grahn A, and Larson G (2013) Targeting the glycoproteome, Glycoconj J 30, 119–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Nilsson J, Ruetschi U, Halim A, Hesse C, Carlsohn E, Brinkmalm G, and Larson G (2009) Enrichment of glycopeptides for glycan structure and attachment site identification, Nat Meth 6, 809–811. [DOI] [PubMed] [Google Scholar]
- [66].Palaniappan KK, and Bertozzi CR (2016) Chemical Glycoproteomics, Chem Rev 116, 14277–14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Boyce M, Carrico IS, Ganguli AS, Yu S, Hangauer MJ, Hubbard SC, Kohler JJ, and Bertozzi CR (2011) Metabolic cross-talk allows labeling of O-linked β-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway, P Natl Acad Sci U S A 108, 3141–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zaro BW, Yang YY, Hang HC, and Pratt MR (2011) Chemical reporters for fluorescent detection and identification of O-GlcNAc-modified proteins reveal glycosylation of the ubiquitin ligase NEDD4–1, P Natl Acad Sci U S A 108, 8146–8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Clark PM, Dweck JF, Mason DE, Hart CR, Buck SB, Peters EC, Agnew BJ, and Hsieh-Wilson LC (2008) Direct in-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins, J Am Chem Soc 130, 11576–11577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Chuh KN, Batt AR, Zaro BW, Darabedian N, Marotta NP, Brennan CK, Amirhekmat A, and Pratt MR (2017) The New Chemical Reporter 6-Alkynyl-6-deoxy-GlcNAc Reveals O-GlcNAc Modification of the Apoptotic Caspases That Can Block the Cleavage/Activation of Caspase-8, J Am Chem Soc 139, 7872–7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Chuh KN, Zaro BW, Piller F, Piller V, and Pratt MR (2014) Changes in metabolic chemical reporter structure yield a selective probe of O-GlcNAc modification, J Am Chem Soc 136, 12283–12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Hao Y, Fan X, Shi Y, Zhang C, Sun D. e., Qin K, Qin W, Zhou W, and Chen X (2019) Next-generation unnatural monosaccharides reveal that ESRRB O-GlcNAcylation regulates pluripotency of mouse embryonic stem cells, Nat Commun 10, 4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Qin W, Qin K, Fan X, Peng L, Hong W, Zhu Y, Lv P, Du Y, Huang R, Han M, Cheng B, Liu Y, Zhou W, Wang C, and Chen X (2018) Artificial Cysteine S-Glycosylation Induced by Per-O-Acetylated Unnatural Monosaccharides during Metabolic Glycan Labeling, Angew Chem Int Ed 57, 1817–1820. [DOI] [PubMed] [Google Scholar]
- [74].Khidekel N, Arndt S, Lamarre-Vincent N, Lippert A, Poulin-Kerstien KG, Ramakrishnan B, Qasba PK, and Hsieh-Wilson LC (2003) A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc posttranslational modifications, J Am Chem Soc 125, 16162–16163. [DOI] [PubMed] [Google Scholar]
- [75].Thompson JW, Griffin ME, and Hsieh-Wilson LC (2018) Chapter Four - Methods for the Detection, Study, and Dynamic Profiling of O-GlcNAc Glycosylation, In Methods in Enzymology (Imperiali B, Ed.), pp 101–135, Academic Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Darabedian N, Thompson JW, Chuh KN, Hsieh-Wilson LC, and Pratt MR (2018) Optimization of Chemoenzymatic Mass Tagging by Strain-Promoted Cycloaddition (SPAAC) for the Determination of O-GlcNAc Stoichiometry by Western Blotting, Biochemistry 57, 5769–5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Greis KD, Hayes BK, Comer FI, Kirk M, Barnes S, Lowary TL, and Hart GW (1996) Selective detection and site-analysis of O-GlcNAc-modified glycopeptides by beta-elimination and tandem electrospray mass spectrometry, Anal Biochem 234, 38–49. [DOI] [PubMed] [Google Scholar]
- [78].Woo CM, Felix A, Byrd WE, Zuegel DK, Ishihara M, Azadi P, Iavarone AT, Pitteri SJ, and Bertozzi CR (2017) Development of IsoTaG, a Chemical Glycoproteomics Technique for Profiling Intact N- and O-Glycopeptides from Whole Cell Proteomes, J Proteome Res 16, 1706–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Hahne H, Sobotzki N, Nyberg T, Helm D, Borodkin VS, van Aalten DM, Agnew B, and Kuster B (2013) Proteome wide purification and identification of O-GlcNAc-modified proteins using click chemistry and mass spectrometry, J Proteome Res 12, 927–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Rexach JE, Rogers CJ, Yu S-H, Tao J, Sun YE, and Hsieh-Wilson LC (2010) Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags, Nat Chem Biol 6, 645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Wang Z, Gucek M, and Hart GW (2008) Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc, Proc Natl Acad Sci U S A 105, 13793–13798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Lefebvre T, Alonso C, Mahboub S, Dupire MJ, Zanetta JP, Caillet-Boudin ML, and Michalski JC (1999) Effect of okadaic acid on O-linked N-acetylglucosamine levels in a neuroblastoma cell line, Biochim Biophys Acta 1472, 71–81. [DOI] [PubMed] [Google Scholar]
- [83].Griffith LS, and Schmitz B (1999) O-linked N-acetylglucosamine levels in cerebellar neurons respond reciprocally to pertubations of phosphorylation, Eur J Biochem 262, 824–831. [DOI] [PubMed] [Google Scholar]
- [84].Woo CM, Lund PJ, Huang AC, Davis MM, Bertozzi CR, and Pitteri SJ (2018) Mapping and Quantification of Over 2000 O-linked Glycopeptides in Activated Human T Cells with Isotope-Targeted Glycoproteomics (Isotag), Mol Cell Proteomics 17, 764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Trinidad JC, Barkan DT, Gulledge BF, Thalhammer A, Sali A, Schoepfer R, and Burlingame AL (2012) Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse, Mol Cell Proteomics 11, 215–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Dias WB, Cheung WD, Wang Z, and Hart GW (2009) Regulation of calcium/calmodulin-dependent kinase IV by O-GlcNAc modification, J Biol Chem 284, 21327–21337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Tarrant MK, Rho HS, Xie Z, Jiang YL, Gross C, Culhane JC, Yan G, Qian J, Ichikawa Y, Matsuoka T, Zachara N, Etzkorn FA, Hart GW, Jeong JS, Blackshaw S, Zhu H, and Cole PA (2012) Regulation of CK2 by phosphorylation and O-GlcNAcylation revealed by semisynthesis, Nat Chem Biol 8, 262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Slawson C, Lakshmanan T, Knapp S, and Hart GW (2008) A mitotic GlcNAcylation/phosphorylation signaling complex alters the posttranslational state of the cytoskeletal protein vimentin, Mol Biol Cell 19, 4130–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Cheung WD, and Hart GW (2008) AMP-activated protein kinase and p38 MAPK activate O-GlcNAcylation of neuronal proteins during glucose deprivation, J Biol Chem 283, 13009–13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Dias WB, Cheung WD, and Hart GW (2012) O-GlcNAcylation of kinases, Biochem Biophys Res Commun 422, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Trinidad JC, Schoepfer R, Burlingame AL, and Medzihradszky KF (2013) N- and O-glycosylation in the murine synaptosome, Mol Cell Proteomics 12, 3474–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Li J, Li Z, Duan X, Qin K, Dang L, Sun S, Cai L, Hsieh-Wilson LC, Wu L, and Yi W (2019) An Isotope-Coded Photocleavable Probe for Quantitative Profiling of Protein O-GlcNAcylation, ACS Chem Biol 14, 4–10. [DOI] [PubMed] [Google Scholar]
- [93].Alfaro JF, Gong CX, Monroe ME, Aldrich JT, Clauss TR, Purvine SO, Wang Z, Camp DG 2nd, Shabanowitz J, Stanley P, Hart GW, Hunt DF, Yang F, and Smith RD (2012) Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets, P Natl Acad Sci U S A 109, 7280–7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Qin W, Lv P, Fan X, Quan B, Zhu Y, Qin K, Chen Y, Wang C, and Chen X (2017) Quantitative time-resolved chemoproteomics reveals that stable O-GlcNAc regulates box C/D snoRNP biogenesis, P Natl Acad Sci U S A 114, E6749–E6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Ma J, Liu T, Wei AC, Banerjee P, O’Rourke B, and Hart GW (2015) O-GlcNAcomic Profiling Identifies Widespread O-Linked beta-N-Acetylglucosamine Modification (O-GlcNAcylation) in Oxidative Phosphorylation System Regulating Cardiac Mitochondrial Function, J Biol Chem 290, 29141–29153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Wang X, Yuan ZF, Fan J, Karch KR, Ball LE, Denu JM, and Garcia BA (2016) A Novel Quantitative Mass Spectrometry Platform for Determining Protein O-GlcNAcylation Dynamics, Mol Cell Proteomics 15, 2462–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Wang Z, Udeshi ND, Slawson C, Compton PD, Sakabe K, Cheung WD, Shabanowitz J, Hunt DF, and Hart GW (2010) Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis, Sci Signal 3, ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Xiao H, Chen W, Smeekens JM, and Wu R (2018) An enrichment method based on synergistic and reversible covalent interactions for large-scale analysis of glycoproteins, Nat Commun 9, 1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Xie S, Jin N, Gu J, Shi J, Sun J, Chu D, Zhang L, Dai CL, Gu JH, Gong CX, Iqbal K, and Liu F (2016) O-GlcNAcylation of protein kinase A catalytic subunits enhances its activity: a mechanism linked to learning and memory deficits in Alzheimer’s disease, Aging Cell 15, 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Gandy JC, Rountree AE, and Bijur GN (2006) Akt1 is dynamically modified with O-GlcNAc following treatments with PUGNAc and insulin-like growth factor-1, FEBS Lett 580, 3051–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Whelan SA, Dias WB, Thiruneelakantapillai L, Lane MD, and Hart GW (2010) Regulation of insulin receptor substrate 1 (IRS-1)/AKT kinase-mediated insulin signaling by O-Linked beta-N-acetylglucosamine in 3T3-L1 adipocytes, J Biol Chem 285, 5204–5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Shi J, Gu JH, Dai CL, Gu J, Jin X, Sun J, Iqbal K, Liu F, and Gong CX (2015) O-GlcNAcylation regulates ischemia-induced neuronal apoptosis through AKT signaling, Sci Rep 5, 14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Hart JR, and Vogt PK (2011) Phosphorylation of AKT: a mutational analysis, Oncotarget 2, 467–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Park SY, Ryu J, and Lee W (2005) O-GlcNAc modification on IRS-1 and Akt2 by PUGNAc inhibits their phosphorylation and induces insulin resistance in rat primary adipocytes, Exp Mol Med 37, 220–229. [DOI] [PubMed] [Google Scholar]
- [105].Niu Y, Xia Y, Wang J, and Shi X (2017) O-GlcNAcylation promotes migration and invasion in human ovarian cancer cells via the RhoA/ROCK/MLC pathway, Mol Med Rep 15, 2083–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kaasik K, Kivimae S, Allen JJ, Chalkley RJ, Huang Y, Baer K, Kissel H, Burlingame AL, Shokat KM, Ptacek LJ, and Fu YH (2013) Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock, Cell Metab 17, 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Wang Z, Pandey A, and Hart GW (2007) Dynamic interplay between O-linked N-acetylglucosaminylation and glycogen synthase kinase-3-dependent phosphorylation, Mol Cell Proteomics 6, 1365–1379. [DOI] [PubMed] [Google Scholar]
- [108].Kazemi Z, Chang H, Haserodt S, McKen C, and Zachara NE (2010) O-linked beta-N-acetylglucosamine (O-GlcNAc) regulates stress-induced heat shock protein expression in a GSK-3beta-dependent manner, J Biol Chem 285, 39096–39107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Qiu H, Liu F, Tao T, Zhang D, Liu X, Zhu G, Xu Z, Ni R, and Shen A (2017) Modification of p27 with O-linked N-acetylglucosamine regulates cell proliferation in hepatocellular carcinoma, Mol Carcinog 56, 258–271. [DOI] [PubMed] [Google Scholar]
- [110].Pyo KE, Kim CR, Lee M, Kim JS, Kim KI, and Baek SH (2018) ULK1 O-GlcNAcylation Is Crucial for Activating VPS34 via ATG14L during Autophagy Initiation, Cell Rep 25, 2878–2890 e2874. [DOI] [PubMed] [Google Scholar]
- [111].Bullen JW, Balsbaugh JL, Chanda D, Shabanowitz J, Hunt DF, Neumann D, and Hart GW (2014) Cross-talk between two essential nutrient-sensitive enzymes: O-GlcNAc transferase (OGT) and AMP-activated protein kinase (AMPK), J Biol Chem 289, 10592–10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Boutros T, Chevet E, and Metrakos P (2008) Mitogen-activated protein (MAP) kinase/MAP kinase phosphatase regulation: roles in cell growth, death, and cancer, Pharmacol Rev 60, 261–310. [DOI] [PubMed] [Google Scholar]
- [113].Cross TG, Scheel-Toellner D, Henriquez NV, Deacon E, Salmon M, and Lord JM (2000) Serine/threonine protein kinases and apoptosis, Exp Cell Res 256, 34–41. [DOI] [PubMed] [Google Scholar]
- [114].Bennmann D, Weidemann W, Thate A, Kreuzmann D, and Horstkorte R (2016) Aberrant O-GlcNAcylation disrupts GNE enzyme activity in GNE myopathy, FEBS J 283, 2285–2294. [DOI] [PubMed] [Google Scholar]
- [115].Chaiyawat P, Chokchaichamnankit D, Lirdprapamongkol K, Srisomsap C, Svasti J, and Champattanachai V (2015) Alteration of O-GlcNAcylation affects serine phosphorylation and regulates gene expression and activity of pyruvate kinase M2 in colorectal cancer cells, Onco Rep 34, 1933–1942. [DOI] [PubMed] [Google Scholar]
- [116].Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard III WA, Peters EC, Driggers EM, and Hsieh-Wilson LC (2012) Phosphofructokinase 1 Glycosylation Regulates Cell Growth and Metabolism Science 337, 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Wells L, Kreppel LK, Comer FI, Wadzinski BE, and Hart GW (2004) O-GlcNAc transferase is in a functional complex with protein phosphatase 1 catalytic subunits, J Biol Chem 279, 38466–38470. [DOI] [PubMed] [Google Scholar]
- [118].Sager RA, Woodford MR, Backe SJ, Makedon AM, Baker-Williams AJ, DiGregorio BT, Loiselle DR, Haystead TA, Zachara NE, Prodromou C, Bourboulia D, Schmidt LS, Linehan WM, Bratslavsky G, and Mollapour M (2019) Post-translational Regulation of FNIP1 Creates a Rheostat for the Molecular Chaperone Hsp90, Cell Rep 26, 1344–1356 e1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Vicart A, Lefebvre T, Imbert J, Fernandez A, and Kahn-Perles B (2006) Increased chromatin association of Sp1 in interphase cells by PP2A-mediated dephosphorylations, J Mol Biol 364, 897–908. [DOI] [PubMed] [Google Scholar]
- [120].Zhao Y, Tang Z, Shen A, Tao T, Wan C, Zhu X, Huang J, Zhang W, Xia N, Wang S, Cui S, and Zhang D (2015) The Role of PTP1B O-GlcNAcylation in Hepatic Insulin Resistance, Int J Mol Sci 16, 22856–22869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Koo J, and Bahk YY (2014) In vivo putative O-GlcNAcylation of human SCP1 and evidence for possible role of its N-terminal disordered structure, BMB Rep 47, 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Ma J, Liu T, Wei AC, Banerjee P, O’Rourke B, and Hart GW (2015) O-GlcNAcomic Profiling Identifies Widespread O-GlcNAcylation in Oxidative Phosphorylation System Regulating Cardiac Mitochondrial Function, J Biol Chem 290, 29141–29153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Kawauchi K, Araki K, Tobiume K, and Tanaka N (2009) Loss of p53 enhances catalytic activity of IKKbeta through O-linked beta-N-acetyl glucosamine modification, P Natl Acad Sci U S A 106, 3431–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Miura T, Kume M, Kawamura T, Yamamoto K, Hamakubo T, and Nishihara S (2018) O-GlcNAc on PKCzeta Inhibits the FGF4-PKCzeta-MEK-ERK1/2 Pathway via Inhibition of PKCzeta Phosphorylation in Mouse Embryonic Stem Cells, Stem Cell Rep 10, 272–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Ning X, Tao T, Shen J, Ji Y, Xie L, Wang H, Liu N, Xu X, Sun C, Zhang D, Shen A, and Ke K (2017) The O-GlcNAc Modification of CDK5 Involved in Neuronal Apoptosis Following In Vitro Intracerebral Hemorrhage, Cell Mol Neurobiol 37, 527–536. [DOI] [PMC free article] [PubMed] [Google Scholar]


