Fig. 3.
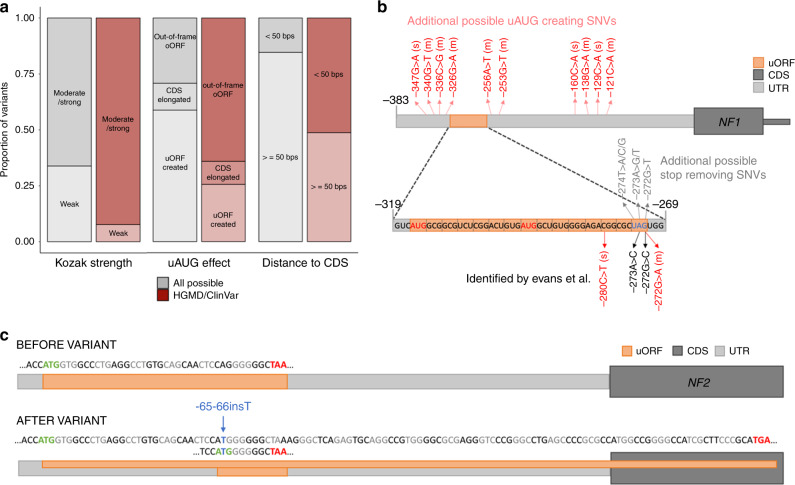
The role of uAUG-creating and uORF stop-removing variants in disease. a The proportion of 39 uAUG variants observed in HGMD and ClinVar (red bars) that fit into different sub-categories compared to all possible uAUG-creating SNVs (grey bars) in the same genes (n = 1022). Compared to all possible uAUG-creating variants, uAUG-creating variants observed in HGMD/ClinVar were significantly more likely to be created into a moderate or strong Kozak consensus (binomial P = 3.5 × 10−4), create an out-of-frame oORF (binomial P = 1.1 × 10−5), and be within 50 bp of the CDS (binomial P = 3.9 × 10−7). b Schematic of the NF1 5’UTR (light grey) showing the location of an existing uORF (orange) and the location of variants previously identified in patients with neurofibromatosis29 in dark red (uAUG-creating) and black (stop-removing). uAUG-creating variants are annotated with the strength of the surrounding Kozak consensus in brackets (“s” for strong and “m” for moderate). All four published variants result in formation of an oORF out-of-frame with the CDS. Also annotated are the positions of all other possible uAUG-creating variants (light red; strong and moderate Kozak only), and stop-removing variants (grey) that would also create an out-of-frame oORF. c Schematic of the NF2 5’UTR (grey) showing the effects of the −65-66insT variant. The reference 5′UTR contains a uORF with a strong Kozak start site. Although the single-base insertion creates a novel uAUG which could be a new uORF start site, it also changes the frame of the existing uORF, so that it overlaps the CDS out-of-frame (forms an oORF). We predict this is the most likely mechanism of pathogenicity. CDS coding sequence, uORF upstream open reading frame, oORF overlapping open reading frame, HGMD the human gene mutation database