Fig. 4.
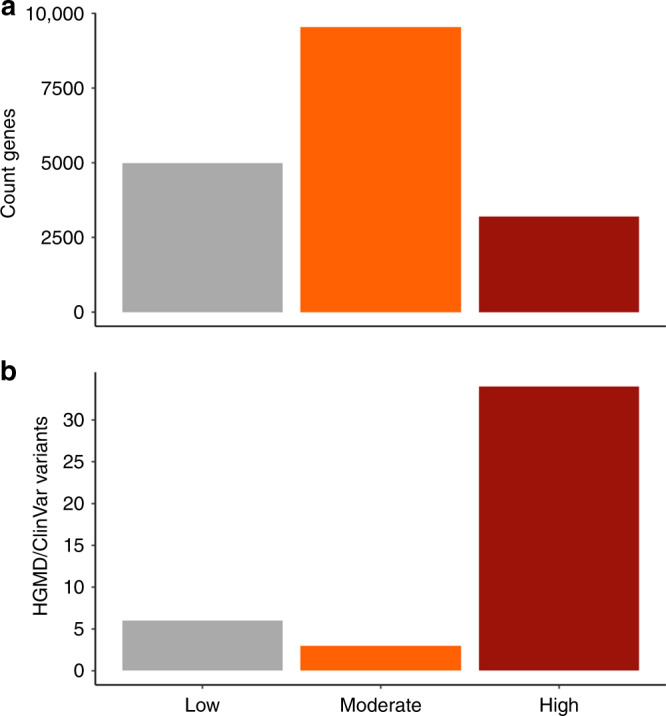
Identifying genes where uORF creating or disrupting variants are likely to have a role in disease. Genes were split into three distinct categories representing a ‘low’, ‘moderate’ and ‘high’ likelihood that uORF-perturbing variants are important. Low likelihood genes include those with existing oORFs, common (>0.1%) oORF creating variants in gnomAD or that are tolerant to LoF. Those in the high likelihood category are remaining genes that are LoF-intolerant or where haploinsufficient or LoF is a known disease mechanism (see methods). a The number of genes in each of the three categories. b The number of uAUG-creating and uORF stop-removing variants in HGMD upstream of genes in each category. Although only 19.2% of all classified genes fall into the high likelihood category (21.4% of all UTR bases when adjusting for UTR length), 83.7% of uORF-perturbing variants identified in HGMD and ClinVar are found upstream of these genes (Fisher’s P = 1.4 × 10−19)
