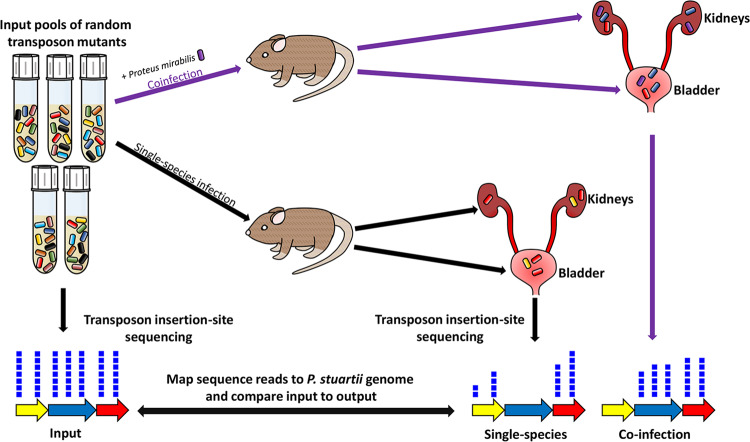
FIG 1.
Conceptual model of single-species and polymicrobial CAUTI Tn-Seq. For each of five transposon mutant library pools, mice were infected as follows using a model of infection that includes the presence of a catheter segment within the bladder: (i) 5 to 10 CBA/J mice were transurethrally inoculated with 1 × 105 CFU of the transposon library for single-species infection (black lines), and (ii) 5 to 10 CBA/J mice were inoculated with 1 × 105 CFU of a 1:1 mixture of the transposon library and wild-type P. mirabilis HI4320 for coinfection (purple lines). Thus, for each input pool, the single-species infections and coinfections were conducted in parallel to utilize the same input inoculum. Input and output samples were enriched for transposon-containing sequences and subjected to next-generation Illumina sequencing of the transposon-chromosome junctions. The resulting reads were mapped to the P. stuartii BE2467 genome, and the abundances of reads at each insertion site from all output samples were compared to those determined for the input samples to determine a fold change value for each gene. The gene indicated in yellow represents a candidate P. stuartii fitness factor for single-species CAUTI that is even more important during coinfection; the gene indicated in blue represents a P. stuartii fitness factor for single-species CAUTI that is no longer important during coinfection; the gene indicated in red represents a factor that does not contribute to P. stuartii CAUTI and was therefore recovered from the infection output pools at a level of density similar to that seen with the input pools.