Figure 1.
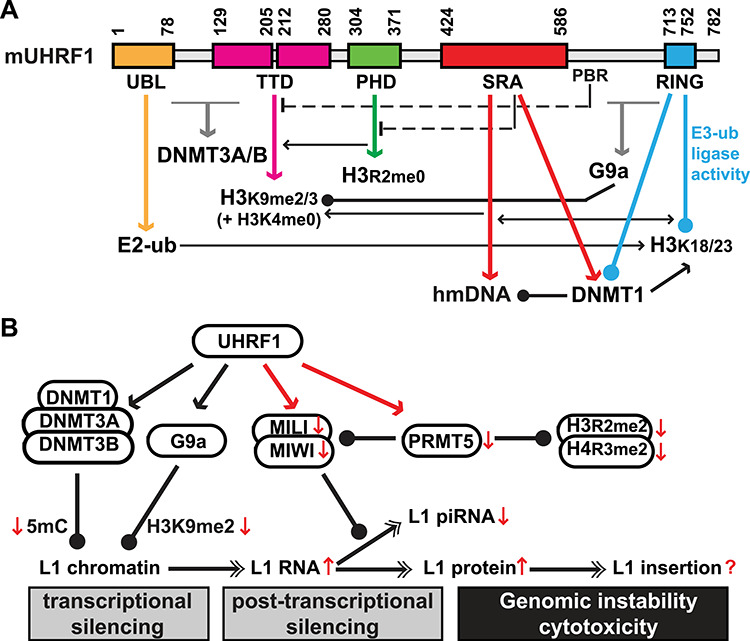
UHRF1 regulates multiple epigenetic pathways during spermatogenesis. (A) Mouse UHRF1 domains and interacting partners. Domain boundaries are labeled above the domain structure as amino acid coordinates per UniProt (https://www.uniprot.org/uniprot/Q8VDF2). Drawn to scale. Six domains/regions are labeled below the domain structure: UBL, ubiquitin like; TTD, tandem Tudor domain; PHD, plant homeodomain; SRA, SET- and RING-associated; PBR, polybasic region; and RING, really interesting new gene. Domains and corresponding interactions (down arrows) are color coded. Interactions with DNMT3A/3B and G9a have not been mapped to defined domains, thus in gray arrows. Ball-headed lines indicate enzymatic modifications. Crosstalk is annotated as either stimulatory (arrow) or inhibitory (T-end dashed lines for intramolecular interactions between TTD and PBR, and between PHD and SRA). hmDNA, hemi-methylated DNA. E2-ub, E2 ubiquitin conjugating enzyme. (B) Model for UHRF1-mediated epigenetic silencing of L1 during spermatogenesis. In postnatal testes, L1 retrotransposons are transcriptionally silenced by DNA methylation and H3K9me2/3, and post-transcriptionally by the pachytene piRNA pathway. In the light of the recent work by Dong and colleagues [4], UHRF1 has emerged as the master regulator of these epigenetic pathways. Newly reported interactions of UHRF1 with PIWI proteins and PRMT5 are highlighted (in red). The conditional loss of UHRF1 function causes meiotic catastrophe and germ cell death. Many changes are observed at molecular levels (↑, upregulation; ↓, downregulation). In particular, L1s are de-repressed at both RNA and protein levels. However, whether there is an increase in L1 insertion and to which degree L1 retrotransposition contributes to genomic instability in Uhrf1-deficient germ cells remain unknown.
