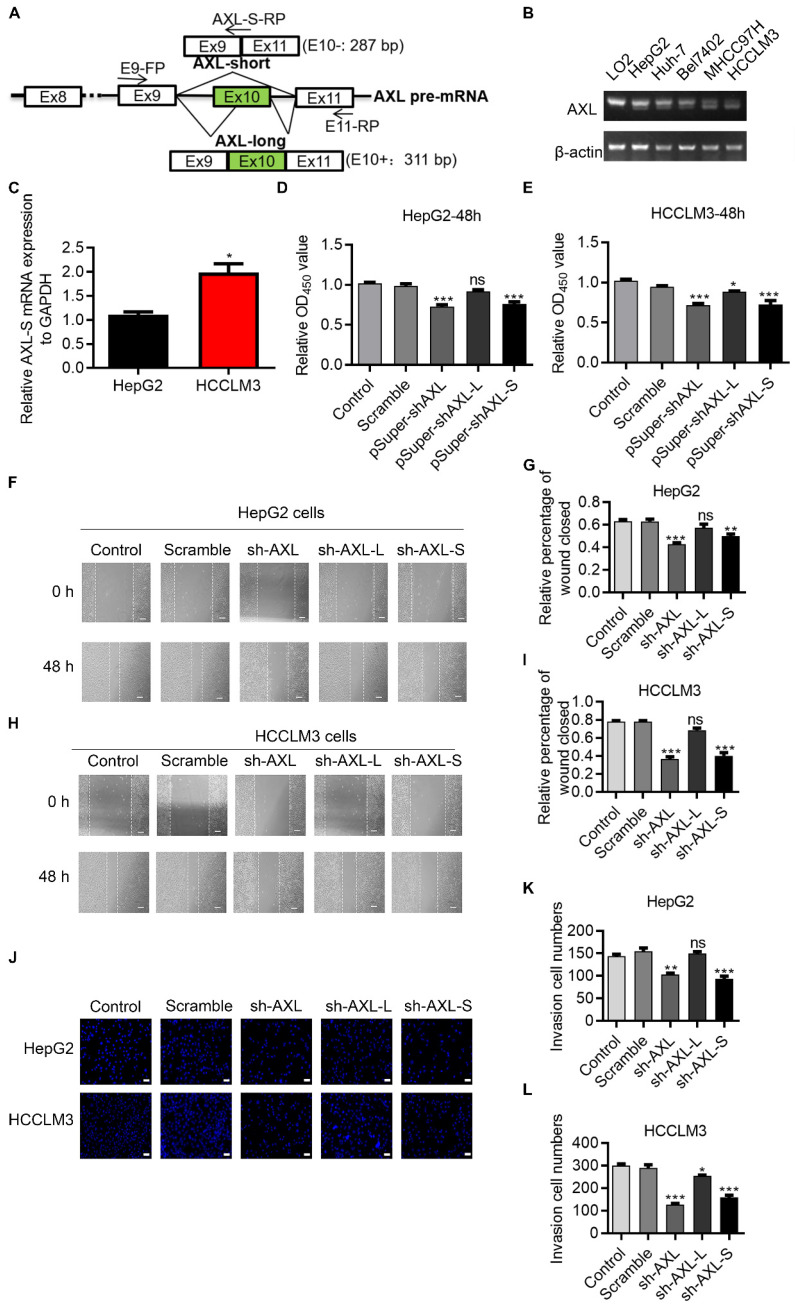
Figure 1.
Oncogenic role of Axl-S isoform in human liver cancer cells. (A) Illustration of Axl pre-mRNA and Axl long and short isoforms; the arrow indicates the design site for the primer. Half of the Axl-S-RP sequence is located in exon 9 and half in exon 11. (B) RT-PCR was used to detect the difference in expression of Axl isoforms between HepG2 and HCCLM3. The primers used were Axl-E9-FP and Axl-E11-RP. The above panel shows the relative expression levels of Axl-S isoform in LO2, HepG2, Huh-7, Bel7402, MHCC97H and HCCLM3. (C) qPCR was used to detect the difference in expression of Axl-S isoform between HepG2 and HCCLM3. The primers used for qPCR detection of Axl-S isoform were Axl-E9-FP and Axl-S-RP. (D-E) The CCK8 assay was used to determine the 48h proliferation of HepG2 and HCCLM3 cells in different groups. The control levels were set to a value of 1. (F-G) Wound healing experiments were performed to examine the effects of knockdown of Axl, Axl-L, or Axl-S on the migration of HepG2 liver cancer cells. (H-I) Wound healing experiments were performed to examine the effects of knockdown of Axl, Axl-L, or Axl-S on the migration of HCCLM3 liver cancer cells. The magnification is 100× and the scale length is 100 μm. (J) The effect of Axl, Axl-L, or Axl-S knockdown on the invasive ability of HepG2 and HCCLM3 liver cancer cells, examined by Transwell assay. The magnification is 200× and the scale length is 50 μm. (K-L) One-way ANOVA and Tukey's post-hoc test were conducted to analyze the invasion HepG2 or HCCLM3. Data are presented as mean ± S.D. (N=3). The “*, **, ***” indicate “P<0.05, 0.01, 0.001” versus the control group, respectively.