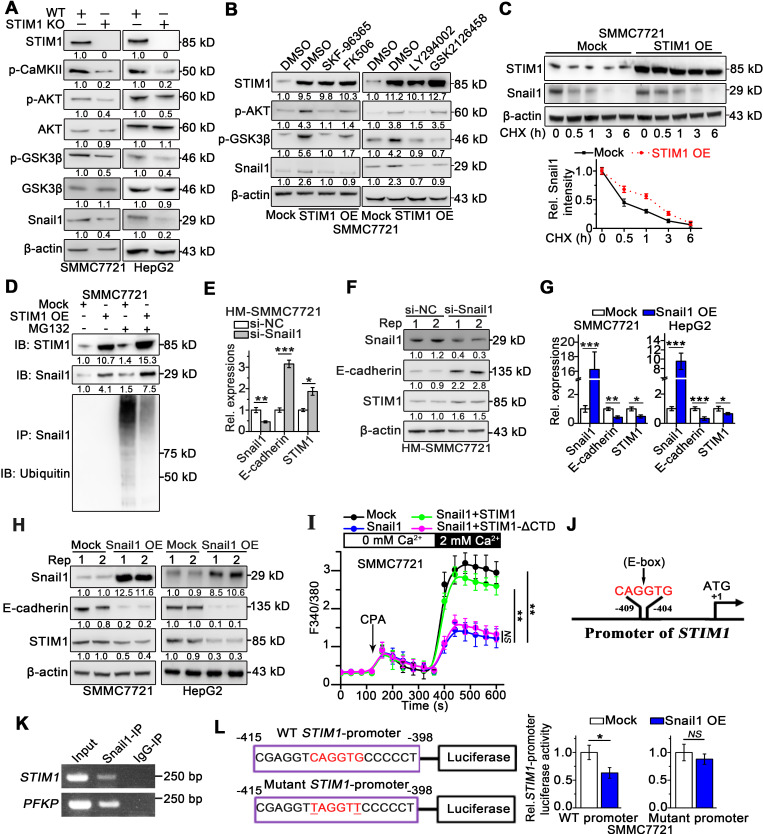
Figure 3.
The interaction between STIM1 and Snail1 in HCC. (A) Indicated protein expressions in WT- and STIM1 KO-SMMC7721 or HepG2 were examined, and β-actin was used as a loading control. (B) STIM1 OE-SMMC7721 cells were treated with SKF-96365 (10 µM), FK506 (10 µM), LY294002 (10 µM), GSK2126458 (1 µM) for 24 h; WB was used for measuring STIM1, p-AKT (Thr308), p-GSK-3β (Ser9) and Snail1 protein levels, and β-actin was used as a loading control, DMSO: dimethyl sulfoxide. (C) Mock- and STIM1 OE-SMMC7721 cells were treated by cycloheximide (CHX, 1 µM) with different time intervals. Cell extracts were immunoblotted with antibodies against STIM1, Snail1 and β-actin. Snail1 levels (normalized to β-actin) were plotted against CHX treatment durations. (D) Mock- and STIM1 OE-SMMC7721 cells were treated with or without MG132 (5 μM). Cell extracts were immunoprecipitated with Snail1 antibody and immunoblotted with antibodies against ubiquitin or Snail1, IP: immunoprecipitation, IB: immunoblotting. (E, F) HM-SMMC7721 sublines were transfected with scrambled siRNA (si-NC) or si-Snail1, RT-qPCR (E) and WB (F) to assess STIM1, Snail1 and E-cadherin expressions. (G, H) RT-qPCR (G) and WB (H) to assess STIM1, Snail1 and E-cadherin expressions in mock- and Snail1 OE-SMMC7721 and HepG2 cells. (I) Ca2+ mobilization upon CPA (20 mM) challenge after over-expressing Snail1, Snail1 plus STIM1, Snail1 plus STIM1-ΔCTD in SMMC7721 cells, respectively, mean ± SEM of 8 independent cells. (J) Bioinformatics analysis predicted binding site of Snail1 (5'-CAGGTG-3') in the promoter of STIM1, black arrow points to transcription start site. (K) ChIP assay of Snail1 protein and STIM1 promoter, representative agarose gel results showing recruitment of Snail1 to the STIM1 promoter, and PFKP promoter used as a positive control. (L) Luciferase activity assay of STIM1 promoter and STIM1 promoter containing mutant E-box (TAGGTT) in Snail1 OE-SMMC7721. Data are expressed as mean ± SEM (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001, NS represents no significant difference.