Extended Data Figure 7. DGAT1 sequence alignment.
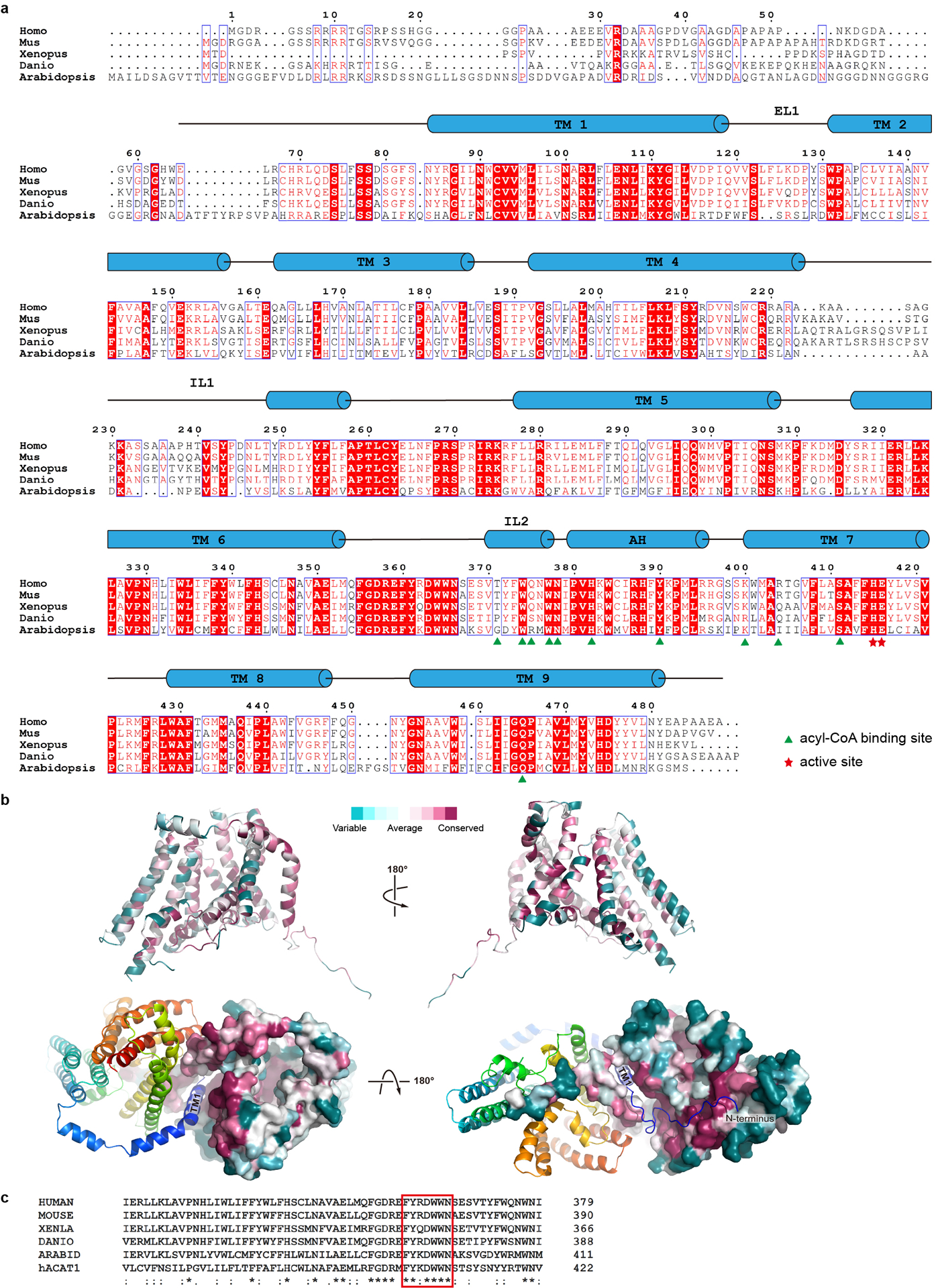
a. DGAT1 of human (Homo, Uniprot accession number O75907), mouse (Mus, Q9Z2A7), frog (Xenopus, A0A1L8G0L4), fish (Danio, Q6P3J0), and thale cress (Arabidopsis, Q9SLD2) are aligned using the Clustal Omega server44. Secondary structural elements of hDGAT1 are marked above the alignment. Residues are colored based on their conservation using the ESPript server45. Residues at the acyl-CoA binding site are marked with green triangles and those at the active site with red stars. b. Consurf mapped onto hDGAT1 structure. hDGAT1 is shown in different orientations in cartoon (top) and surface (bottom) representations. One hDGAT1 monomer is colored based on the conservation score of each residue, calculated by the Consurf server46. c. Sequence alignments of the FYXDWWN motif (red rectangle) from DGAT1 of different species and human ACAT1 (Uniprot accession number: P35610).
