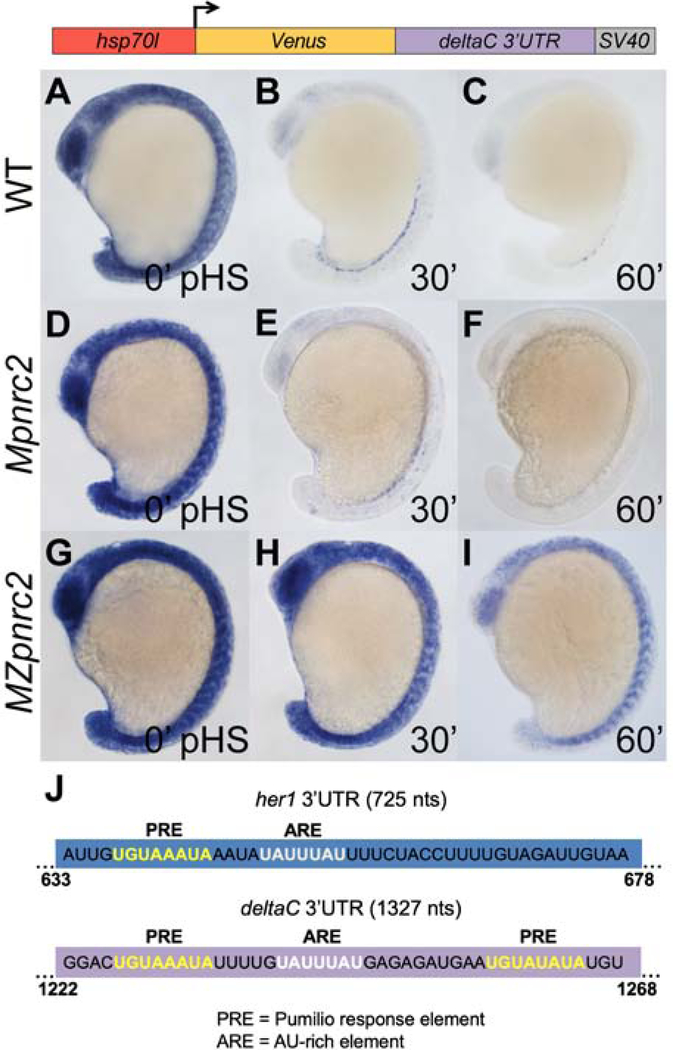
Figure 5. The dlc 3’UTR confers Pnrc2-mediated decay to reporter transcripts.
(A–C) Transgenic embryos carrying the hsp70l:Venus-dlc 3’UTR-SV40 pA reporter (line oz81) were raised to mid-segmentation stage and heat shocked for 15 minutes, then collected at the indicated minutes pHS and processed by Venus in situ hybridization (n ≥ 6 embryos per time point). Venus transcript is not detected in the absence of heat shock (n = 10 embryos) (data not shown). (D–I) Mpnrc2 (D–F) and MZpnrc2 mutant embryos (G–I) carrying the hsp70l:Venus-dlc 3’UTR-SV40 pA reporter (line oz81) were also heat shocked and processed for Venus transcript (n ≥ 10 embryos per time point). Representative embryos were genotyped post-imaging to confirm genotype. (J) RBPmap motif analysis (Paz et al., 2014) identifies a canonical PRE (5’UGUAAAUA; yellow) and a canonical ARE (5’UAUUUAU; white) near the end of the her1 3’UTR and two PREs and one ARE near the end of the dlc 3’UTR. The indicated PRE and ARE are the only such motifs in the 725 nt full-length her1 3’UTR, whereas the 1327 nt dlc 3’UTR contains an additional PRE and two additional AREs. Three independent lines carrying the hsp70l:Venus-dlc 3’UTR-SV40 pA reporter were analyzed in wild-type embryos by in situ hybridization and exhibited comparable Venus decay across all three lines (data not shown); one representative line is shown (see Methods for details). pHS = post-heat shock; nts = nucleotides; PRE = Pumilio response element; ARE = AU-rich element; pA = polyadenylation sequence.