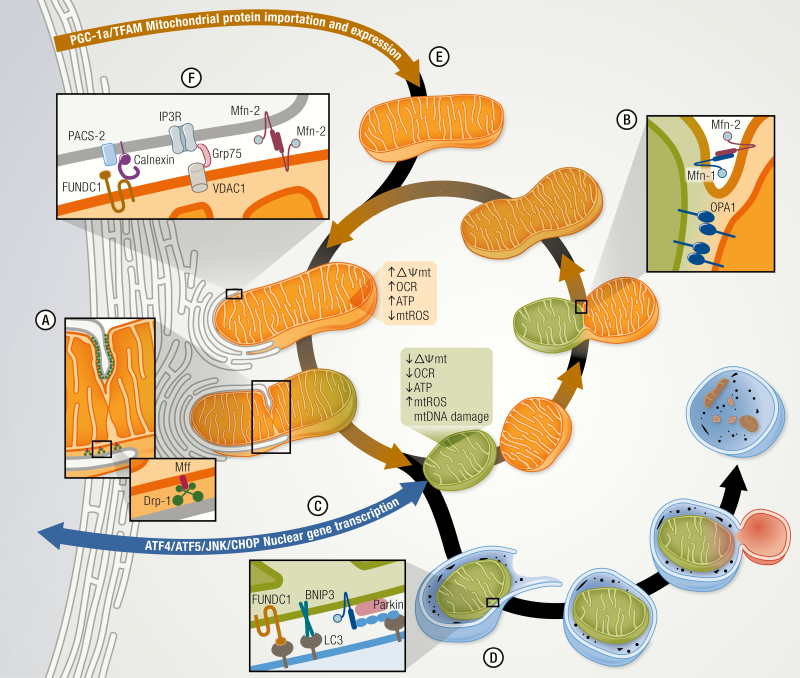
Figure 1.
Mitochondrial plasticity regulates the mitochondrial quality control and function. A mitochondrion is a malleable organelle that constantly changes its structure and shape to adapt its function and allow the normal replacement of anomalous mitochondria. (A) Mitochondrial fission is regulated by Drp-1, which binds to outer mitochondrial membrane (OMM) protein adaptor like Mff, which together with the endoplasmic reticulum, (ER) constrict and isolate the impaired mitochondria. On the other hand, mitochondrial fusion (B) is orchestrated by dynamin-related GTPases mitofusin (Mfn) 1/2 and optic atrophy type 1 (OPA1) promoting the OMM and inner mitochondrial membrane (IMM) fusion respectively. This process allows the function of an anomalous mitochondrion to be enhanced and averaged through combination with a healthy mitochondrion. (C) The stressed mitochondria also communicate with the nucleus through the mitochondrial unfolded protein response (UPRmt) and activate ATF4/5 and JNK/CHOP signaling pathways to induce the transcription of nuclear genes associated with mitochondrial survival and quality control. (D) Alternatively, impaired mitochondria may also be eliminated by mitophagy, triggered by the direct association of BNIP3 or FUNDC1 with LC3 or by marking mitochondria through the parkin-mediated polyubiquitination of OMM proteins. (E) Nuclear encoded mitochondrial proteins and mtDNA transcription are induced by PGC-1α that enhance mitochondrial biogenesis and maintain the mitochondrial mass. (F) ER–mitochondria communication through mitochondria-associated membranes (MAMs) allow molecules to be exchanged between these organelles and regulate the mitochondrial metabolism, bioenergetics, and signaling processes. MAMs are composed of ER proteins like PACS-2, Calnexin, and Mfn-2 and by mitochondrial proteins like VDAC, Mfn-2, and FUNDC1 among others.