Figure 2.
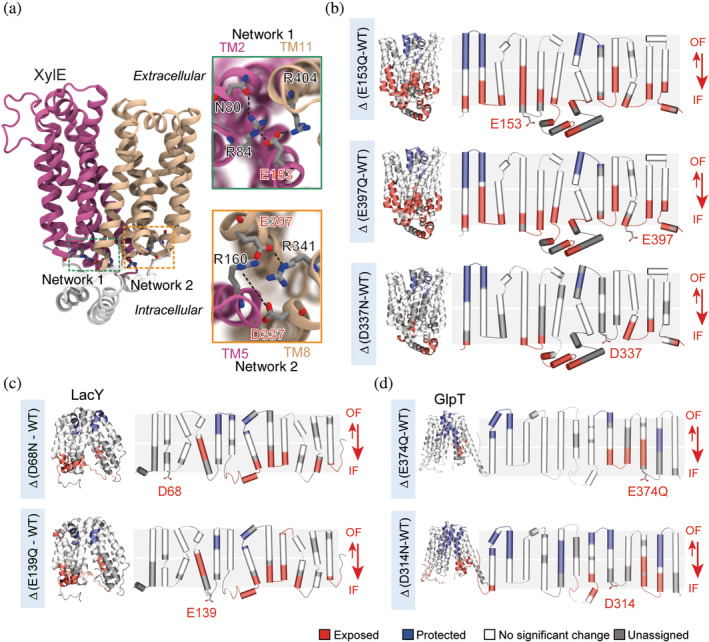
ΔHDX‐MS on MFS transporters LacY, XylE, and GlpT identifies a conserved mechanism of conformational cycling. (a) Two charge‐relay networks of conserved residues stabilize the OF state of XylE (PDB: 4GBY). Close‐up of networks 1 and 2—highlighted in green and orange, respectively—show the connection between the two lobes and highlight in red the mutated acidic residues. (b) ΔHDX between the mutants E153Q, E397Q, and D337N versus WT mapped onto the 3D and topological structure of XylE. (c) ΔHDX of the mutants D68N, and E139Q versus WT mapped onto the 3D and topological structure of LacY. (d) ΔHDX of the mutants E374Q and D314N mapped onto the 3D and topological structure of GlpT. Reproduced with permission from Reference 19 (distributed under CC‐BY license). HDX‐MS, hydrogen–deuterium exchange coupled to mass spectrometry; MFS, major facilitator superfamily; OF, outward facing; PDB, Protein Data Bank coordinates; WT, wild type
